AbstractPurposeNeuregulin 1 (NRG1) gene fusion is a potentially actionable oncogenic driver. The oncoprotein binds to ERBB3-ERBB2 heterodimers and activates downstream signaling, supporting a therapeutic approach for inhibiting ERBB3/ERBB2. However, the frequency and clinicopathological features of solid tumors harboring NRG1 fusions in Korean patients remain largely unknown.
Materials and MethodsWe reviewed archival data from next-generation sequencing panel tests conducted at a single institution, specifically selecting patients with in-frame fusions that preserved the functional domain. The clinicopathological characteristics of patients harboring NRG1 fusions were retrospectively reviewed.
ResultsOut of 8,148 patients, NRG1 fusions were identified in 22 patients (0.27%). The average age of the patients was 59 years (range, 32 to 78 years), and the male-to-female ratio was 1:1.2. The lung was the most frequently observed primary site (n=13), followed by the pancreaticobiliary tract (n=3), gastrointestinal tract (n=2, stomach and rectum each), ovary (n=2), breast (n=1), and soft tissue (n=1). Histologically, all tumors demonstrated adenocarcinoma histology, with the exception of one case of sarcoma. CD74 (n=8) and SLC3A2 (n=4) were the most frequently identified fusion partners. Dominant features included the presence of fewer than three co-occurring genetic alterations, a low tumor mutation burden, and low programmed death-ligand 1 expression. Various clinical responses were observed in patients with NRG1 fusions.
IntroductionSubsets of gene fusions in solid tumors have been identified as oncogenic drivers and potential therapeutic targets [1,2]. Among them, tyrosine kinases such as ALK, ROS1, RET, NTRK, and FGFR are frequently involved in oncogenic fusions. These fusions lead to the constitutive activation of downstream signaling pathways, promoting tumor cell proliferation and tumor progression [2].
Neuregulin 1 (NRG1), which is encoded by the NRG1 gene, functions as an epidermal growth factor (EGF)–like ligand for members of the ERBB/epidermal growth factor receptor (EGFR) family, particularly human epidermal growth factor receptor 3 (HER3/ERBB3) [3]. This interaction results in the formation of heterodimers between HER3 and other ERBB family members, predominantly HER2 [4]. NRG1 fusion was initially identified in the breast cancer cell line MDA-MB-175, where it manifested as a DOC4-NRG1 transcript that was specific to the tumor and played a role in promoting tumor cell proliferation [5,6]. Subsequently, NRG1 fusion has been observed in various cancer types [7,8], with a notably high prevalence in pulmonary invasive mucinous adenocarcinoma (IMA) [9–11].
NRG1 fusion is a rare occurrence, reported to have an incidence of 0.2% based on a study involving 21,858 tumors with CD74 being the most common partner [12]. Despite its rarity, NRG1 fusion in solid tumors is now recognized as a target for drug therapy with evidence indicating a sustained response to targeted agents such as a pan-ErbB family inhibitor (afatinib) [13] and anti-HER2xHER3 bispecific antibody (zenocutuzumab) [14,15]. However, the frequency and clinicopathological features of solid tumors harboring NRG1 fusions, particularly in Korean patients, remain largely unknown. In this study, we aimed to investigate the incidence and clinicopathological characteristics of NRG1 fusion–positive solid tumors through a comprehensive molecular analysis.
Materials and Methods1. PatientsBetween May 2017 and June 2022, a total of 8,148 patients with solid tumors underwent next-generation sequencing (NGS) analysis. Among them, 22 patients who exhibited NRG1 fusions were included in this study. Clinical information and outcome data of the patients, including age at diagnosis, sex, primary tumor site, tumor stage, smoking status, pathological diagnosis, programmed death-ligand 1 (PD-L1) status, and survival data, were retrieved from electronic medical records. The best overall response to systemic therapy was evaluated based on the Response Evaluation Criteria in Solid Tumor ver. 1.1 criteria.
2. Histopathologic reviewAll available pathological slides stained with hematoxylin and eosin (H&E) were reviewed by two pathologists (Y.J.C. and H.S.S.). Their objective was to assess tumor differentiation and histological types. Since the majority of cases involved adenocarcinomas, the architectural features of the tumors were carefully examined, specifically focusing on the presence of tubule/cribriform formation, micropapillary clusters, and papillary configuration. Since previous studies have reported an association between NRG1 fusion and mucinous adenocarcinoma [9,10], we examined the following cytologic features: cell morphology (especially columnar cells), intracellular or extracellular mucin, and cytoplasmic vacuoles. Lung adenocarcinomas were classified into two types based on cytological features and thyroid transcription factor-1 (TTF-1) expression: the terminal respiratory unit (TRU) type and the non-TRU type [16–18].
3. Immunohistochemistry and interpretation of PD-L1Immunohistochemistry (IHC) was conducted using the following procedure. Tissue sections, 4-μm thick, were cut from formalin-fixed paraffin-embedded (FFPE) tissue blocks. The sections were deparaffinized with xylene and rehydrated using graded alcohol solutions. IHC staining was carried out utilizing a Ventana Discovery XT Automated Slide Stainer (Ventana Medical Systems, Tucson, AZ). Antigen retrieval was performed using Conditioning 1 buffer (citrate buffer, pH 6.0, Ventana Medical Systems). The slides were then incubated with primary antibodies against PD-L1 (pre-diluted, clone SP263, Ventana Medical System) and TTF-1 (1:150, 8G7G3/1, Agilent Dako, Santa Clara, CA). For PD-L1 (22C3), the PD-L1 IHC 22C3 pharmDx kit on the Dako Automated Link 48 platform (Dako, Carpenteria, CA) was utilized. Interpretation of the IHC results was conducted by two pathologists (Y.J.C. and H.S.S.). For PD-L1 IHC, the tumor proportion score (TPS) was assessed. TPS represents the percentage of viable tumor cells demonstrating partial or complete membrane staining at any intensity. Positive PD-L1 expression was defined as TPS ≥ 1% using the SP263 or 22C3 IHC assays. TTF-1 expression was considered positive when 10% or more of tumor cells exhibited nuclear staining, as described in previous studies [17,18].
4. NGS analysisTargeted DNA and RNA sequencing was conducted using the TruSight Tumor 170 (TST170, Illumina, San Diego, CA) or TruSight Oncology 500 (TSO500, Illumina) panels. The TST170 panel is designed to identify 170 cancer-related genes, which encompass 151 genes associated with potential single-nucleotide variants (SNVs) and indels, 59 genes associated with potential amplifications, and 55 genes associated with RNA fusions and splice variants. The TSO500 panel comprises 523 cancer-related genes utilized for calculating tumor mutation burden (TMB), 130 regions for assessing microsatellite instability (MSI), and an equal number of genes as TST170 for detecting potential amplification, RNA fusions, and RNA splice variants.
Following the established protocol [19], DNA and RNA were extracted from 40 ng of FFPE tissue using the Qiagen AllPrep DNA/RNA FFPE Kit (Qiagen, Hilden, Germany). Target enrichment was achieved through hybridization capture, and paired-end sequencing (2×150 bp) was conducted using a NextSeq sequencer (Illumina) following the manufacturer’s instructions. In the analysis, variants meeting the following criteria were included: a total depth of at least 100× and a variant allele frequency of at least 3%. The interpretation of variants followed the recommendations provided by the Association for Molecular Pathology, the American Society of Clinical Oncology, and the College of American Pathologists [20].
5. RNA fusion detection and interpretationRNA fusions were detected using the Manta fusion caller [21] integrated into the analysis pipeline of TST170 or TSO500 Local App ver. 2.0 or 2.2, respectively. For validating RNA fusions and interpretation with a visualized plot, Arriba RNA fusion caller (ver. 2.2.1) [22] was used. STAR (ver. 2.7.10a) [23] aligned bam with the option–chimOutType ‘WithinBAM HardClip Junctions.’ The detected fusion plots were generated using the R script draw_fusion.R, which visualizes the fusion partners, breakpoints, supporting reads, fusion type, frame status, and retained protein domains. For NRG1 fusion to be considered functional, it needed to meet the following criteria: (1) predicted in-frame fusion, (2) located at the C-terminal region with a retained EGF-like domain, and (3) supporting split or discordant reads ≥ 5 [24].
6. Tumor mutation burdenThe TMB scores were calculated using TSO500, which covered 1.33 megabases (Mb) coding regions. According to the TSO500 LocalApp manual, we included SNVs or insertions or deletions in the coding region with a variant allele frequency between ≥ 5% and < 90% and a read depth ≥ 50×. However, we excluded certain variants that were annotated with ≥ 50 counts in the COSMIC database, ≥ 10 counts in normal population databases (gnomAD exome/genome, 1000 Genome), or were present in the internal germline variant database.
Results1. Characteristics of patients and prevalence of NRG1 fusionThe clinicopathological characteristics of the patients are presented in Table 1. NRG1 fusion was identified in 22 out of 8,148 patients (0.27%) with solid tumors. Among the NRG1 fusion–positive patients, there were 10 males and 12 females, with a median age of 59 years (range, 39 to 78 years). Of these patients, 15 (68.2%) reported never smoking, while five were former smokers and two were current smokers (median pack-years: 15 and 21.5, respectively). The lung was the most common primary site of NRG1 fusion–positive tumors (n=13, 59.1%). Following that, the pancreaticobiliary (PB) tract (n=3 [2 pancreatic cancer, 1 intrahepatic cholangiocarcinoma], 13.6%), gastrointestinal (GI) tract (n=2 [1 rectal cancer, 1 gastric cancer], 9.1%), ovary (n=2, 9.1%), breast (n=1, 4.6%), and soft tissue (n=1 [dedifferentiated liposarcoma], 4.6%) (Fig. 1). Regarding the primary tumor site, NRG1 fusion was detected in 0.72% (13/1,814) of all lung cancer patients with available NGS reports, followed by breast (0.49%, 1 out of 205) and PB (0.39%, 3 out of 770) cancers (Fig. 2).
2. Pathologic features of NRG1 fusion–positive tumors
Table 2 summarizes the pathological characteristics of the NRG1 fusion–positive tumors. All of the NRG1 fusion–positive tumors had adenocarcinoma histology, except for one case of dedifferentiated liposarcoma with spindle cell morphology (Fig. 3). Among the adenocarcinomas, non-TRU type lung adenocarcinoma, including mucinous adenocarcinoma, was the most common subtype (38.1%), followed by TRU type lung adenocarcinoma (23.8%) and GI and PB adenocarcinoma (23.8%). Regarding architectural features, all adenocarcinoma cases exhibited at least one typical pattern, with micropapillary patterns frequently observed (76.2%). Solid sheet patterns and undifferentiated areas were not prominently present. Among the adenocarcinoma cases, 61.9% displayed cancer cells with columnar cell morphology. Furthermore, the majority of (95.2%) showed at least intracellular mucins or cytoplasmic vacuoles on H&E slides.
PD-L1 IHC was performed on 18 patients. Four patients (18.2%) were PD-L1–positive (three with lung adenocarcinoma and one with ovarian serous carcinoma). No high expression (TPS ≥ 50%) was observed. Eight cases (61.5%) of lung adenocarcinoma (n=13) were TTF-1 IHC-negative.
3. Genomic features of NRG1 fusion–positive tumorsOf the 22 cases, seven (31.8%) were tested using TST170, and the remaining 15 (81.8%) were tested using TSO500. Translocation (n=17, 77.3%) was the most common type of alteration, followed by inversion (n=4, 18.2%) and deletion (n=1, 4.5%). Among the NRG1 fusion cases, the most frequent breakpoints occurred in exon 6 (n=13, 59.1%), and exon 2 (n=8, 36.4%). The most prevalent fusion partner was CD74 (n=8, 36.4%), followed by SLC3A2 (n=3, 13.6%). Both CD74 and SLC3A2 formed fusion transcripts with NRG1 exon 6. The other fusion partners included ATP1B1, CDH1, CLU, CRADD, FUT10, INCENP, KIF22, RBPMS, SLC20A2, VWA8, and XKR6. Seventeen (77.3%) NRG1 fusion–positive tumors had one or more co-occurring mutations: 1 (n=9, 40.9%), 2 (n=2, 9.1%), 3 (n=5, 22.7%), and 6 (n=1, 4.5%). All were EGFR/ALK/ROS1-wild type. TP53 mutation was the most co-occurring mutation, present in 10 cases (54.5%). KRAS (G12S and G12D), BRAF (N581I), and PIK3CA (E453K) mutations were observed concurrently in four cases. TMB and MSI were calculated for 15 cases. The median value of TMB was 3.9/Mb (range, < 1.0 to 51.20/Mb), and the median value of MSI was 1.98% (range, < 1.0 to 5.0%). The genomic findings are summarized in Fig. 4.
4. Clinical outcome of NRG1 fusion–positive tumorsMost patients were in an advanced stage at diagnosis, with nine stage IV patients (40.9%). Among the 13 patients diagnosed with stages I–III, 10 underwent surgical resection, and all patients experienced recurrence or metastasis. One patient with liposarcoma received repeated radiotherapy as the sole treatment, while the remaining patients were treated with systemic chemotherapy. Although it is not appropriate to compare survival across different tumor types, the treatment sequence and survival after recurrence/metastasis in patients with NRG1 fusion are depicted in Fig. 5. Among the 13 patients with lung cancer, four (P02, P10, P20, and P21) received NRG1 fusion–targeting treatment (anti-HER2xHER3 bispecific antibody zenocutuzumab) and were still undergoing treatment at the cut-off date with a durable response. However, one patient with pancreatic cancer (P06) was also treated with NRG1 fusion–targeting treatment after poor responses to the two previous treatment lines but showed short progression-free survival (1 month) and died (overall survival, 7.2 months from initial diagnosis).
DiscussionIn this study, we report 22 cases of NRG1 fusion–positive solid tumors, accounting for 0.27% of the 8,148 cases of solid tumors with available NGS reports reviewed in a single tertiary institution. The study focuses on the clinical, pathological, and genetic characteristics of these cases. NRG1 fusions were detected in various tumors and primary sites, occurring at a rare incidence. The adenocarcinoma histology was predominant among the NRG1 fusion–positive tumors. Most patients presented with advanced-stage disease and showed various responses to systemic treatment.
In this study, the prevalence of NRG1 fusion was 0.27%, consistent with 0.2% in a previous report [12]. All but one patient demonstrated adenocarcinoma histology. This exceptional case exhibited sarcoma, consistent with a previous study that reported one case of sarcoma (0.2% frequency) [12]. In previous studies, NRG1 fusion was most frequently detected in pulmonary IMA [9,12]. IMA has a columnar cell morphology, harboring intracytoplasmic mucin [25]. These findings (columnar cells or intracellular mucins) were observed in > 50% of patients in this cohort. In cases of lung adenocarcinoma, typical IMAs and adenocarcinomas with eosinophilic cytoplasm in columnar cell morphology, previously referred to as a subtype of non-TRU adenocarcinoma [18], were frequently encountered. In this study, all adenocarcinomas with these morphological features tested negative for TTF-1, leading to their collective classification as non-TRU type adenocarcinomas. Remarkably, the non-TRU type accounted for the largest proportion (61.5%) of lung adenocarcinomas, which partly agrees with a previous study where 57% of NRG1 fusion–positive lung tumors were mucinous adenocarcinoma [11]. Despite occurring in the lungs, non-TRU type adenocarcinoma exhibits morphological and molecular characteristics similar to GI adenocarcinoma [26]. Thus, considering the total number of GI-PB adenocarcinomas identified in this study, 61.9% showed a gut phenotype. However, it is worth noting that NRG1 fusion was not exclusively limited to these adenocarcinomas, as it was also detected in TTF-1–positive TRU-type adenocarcinomas or other types of carcinomas without mucin or columnar morphology. Hence, these findings indicate that NRG1 fusion should not be ruled out solely based on histological findings. Interestingly, no solid sheet pattern was observed; however, a micropapillary pattern was present in 76.2% of the cases. Considering the well-established association between the micropapillary pattern and poor prognosis, it is reasonable to assume that this histological feature correlates with the frequent recurrence of NRG1-positive tumors observed in this study.
Of the 17 out of 22 cases with co-occurring mutations, most had three or fewer concurrent mutations. Interestingly, we found two co-occurring KRAS mutations in one lung IMA (G12S) and one PB cancer (G12D). KRAS mutations have been reported to be mutually exclusive with NRG1 fusions in pancreatic cancer [27]. However, concurrent KRAS or EGFR mutations have been reported in IMAs harboring NRG1 fusion [11]. In a preclinical study using lung cancer cells, the coexistence of KRAS mutation and NRG1 fusion synergistically activated tumor cell proliferation and conferred treatment resistance [28]. In this study, we observed a poor prognosis in a patient with PB carcinoma harboring co-occurring KRAS mutations.
All the patients showed no or low PD-L1 expression. No patient exhibited high PD-L1 expression. In addition, all cases, except for two, had a low TMB (less than 10/Mb). These findings suggest that NRG1 fusion–positive tumors exhibit lower response rates to immune checkpoint inhibitors. The lower response rate observed in the eNRGy1 patient cohort supports this notion [11]. This indicates that alternative treatment strategies need to be explored for NRG1 fusion–positive tumors to improve therapeutic outcomes.
Targeting the HER2/HER3 pathway is a treatment approach for NRG1 fusion–positive tumors. Zenocutuzumab (Zeno, MCLA-128) is a bispecific HER2/HER3 antibody with enhanced antibody-dependent cytotoxicity. Zenocutuzumab binds to HER2 and interferes with the interaction between NRG1 and HER3, inhibiting HER2/HER3 heterodimerization [14,15,29]. Recently, a global multicenter phase I/II trial (the eNRGy trial) investigated the clinical efficacy and safety of the anti-HER2xHER3 bispecific antibody zenocutuzumab for NRG1 fusion–positive solid tumors and reported durable efficacy [30]. In our institution, four patients with lung cancer were treated with zenocutuzumab in the eNRGy trial and showed good responses and long treatment durations. However, as previously mentioned, one patient with pancreatic cancer did not respond to zenocutuzumab. Further trials are needed to validate the efficacy and safety of anti-NRG1 treatment in different tumor types.
This study had several limitations. Because of the rarity of NRG1 fusion, although we examined over 8,000 tumors with NGS, only 22 cases were reviewed in the study, which might be insufficient to perform an appropriate statistical analysis. Except for the lungs, NRG1 fusion was detected in fewer than three cases at each primary site, which is too few to determine the exact prevalence of different populations of tumors at different sites. As many of the specimens used to observe the histological findings were biopsy specimens, the morphological findings described above may not represent their entirety. In addition, most patients included in this study were treated with heterogeneous conventional systemic therapy depending on the primary site and tumor stage, making the appropriate evaluation of clinical outcomes complex and difficult.
In conclusion, we conducted a collective analysis of NRG1 fusion–positive tumors at a single institution. NRG1 fusion is a rare occurrence, and patients are frequently diagnosed at an advanced stage, predominantly with adenocarcinoma, showing diverse responses to therapeutic regimens. Despite its rarity, the detection of NRG1 fusions holds significance as novel targeted agents are being developed. In patients with advanced-stage adenocarcinoma, NGS could serve as a valuable tool for screening various oncogenic driver gene alterations, including NRG1 fusion.
NotesEthical Statement This study was approved by the Institutional Review Board (local IRB number: 4-2022-1165). As this was a retrospective study, the IRB waived the requirement for informed consent due to the study design. Author Contributions Conceived and designed the analysis: Cha YJ, Lee C, Lee CK, Shim HS. Collected the data: Cha YJ, Lee C, Joo B, Kim KA, Lee CK, Shim HS. Contributed data or analysis tools: Cha YJ, Lee C, Joo B, Kim KA, Lee CK, Shim HS. Performed the analysis: Cha YJ, Lee C, Joo B, Kim KA, Lee CK, Shim HS. Wrote the paper: Cha YJ, Lee C, Lee CK, Shim HS. AcknowledgmentsThis work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT) (No.2022R1A2C1009364).
Fig. 2Detection rate of neuregulin 1 (NRG1) fusion in all patients and in different primary sites. Etc (n=170) includes thyroid (n=50), thymus (n=38), lymphoma (n=28), malignancy of unknown primary (n=21), skin (n=17), testis (n=13), and adrenal gland (n=3). 
Fig. 3Histologic features of neuregulin 1 (NRG1) fusion–positive tumors. (A) Typical histology of invasive mucinous adenocarcinoma (H&E, ×200). (B) Columnar tumor cells with intraluminal necrosis (H&E, ×200). (C) Columnar tumor cells in papillary architecture (H&E, ×200). (D) Micropapillary tumor clusters floating in the extracellular mucin pool (H&E, ×200). (E) Adenocarcinoma with tubular formation (H&E, ×200). 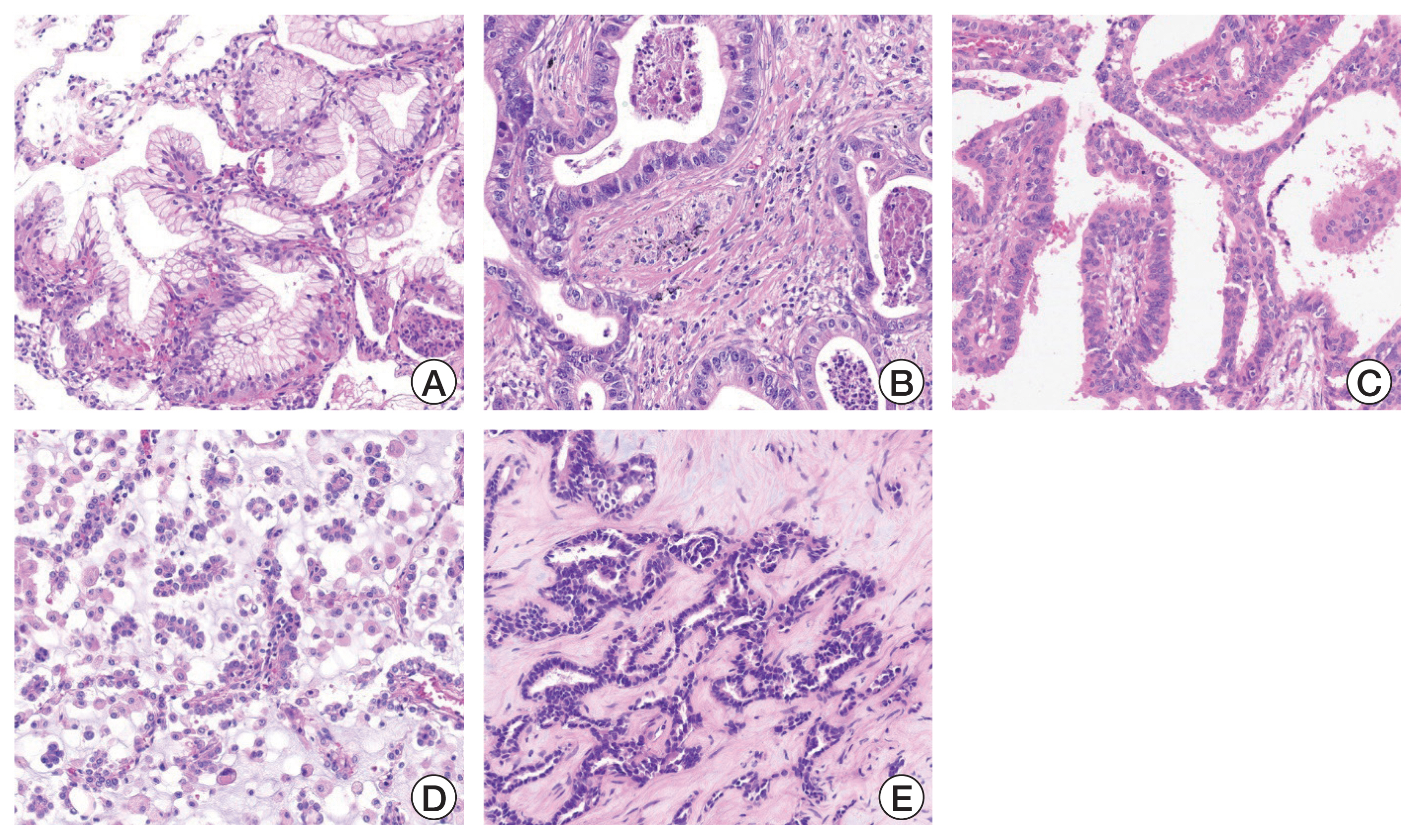
Fig. 4Summary of co-occurring genomic alterations. CNV, copy number variation; GI, gastrointestinal; INDEL, insertion and deletion; NA, not available; PB, pancreaticobiliary; SNV, single-nucleotide variant; TTF-1, thyroid transcription factor-1. 
Fig. 5Swimmer plot displaying treatment sequence and survival of patients with neuregulin 1 (NRG1) fusion. Swimmer plot showing treatment sequences (first-line and up to fourth-line of systemic treatment) with survival from diagnosis of recurrence/metastasis until the last follow-up. NRG1 fusion partner genes and primary tumor sites are shown. The duration of NRG1 targeting treatment is also shown. Four patients with lung cancer (P02, P10, P20, and P21) are undergoing NRG1 targeting treatment at the cut-off date with a durable response. GI, gastrointestinal tract; PB, pancreaticobiliary tract. 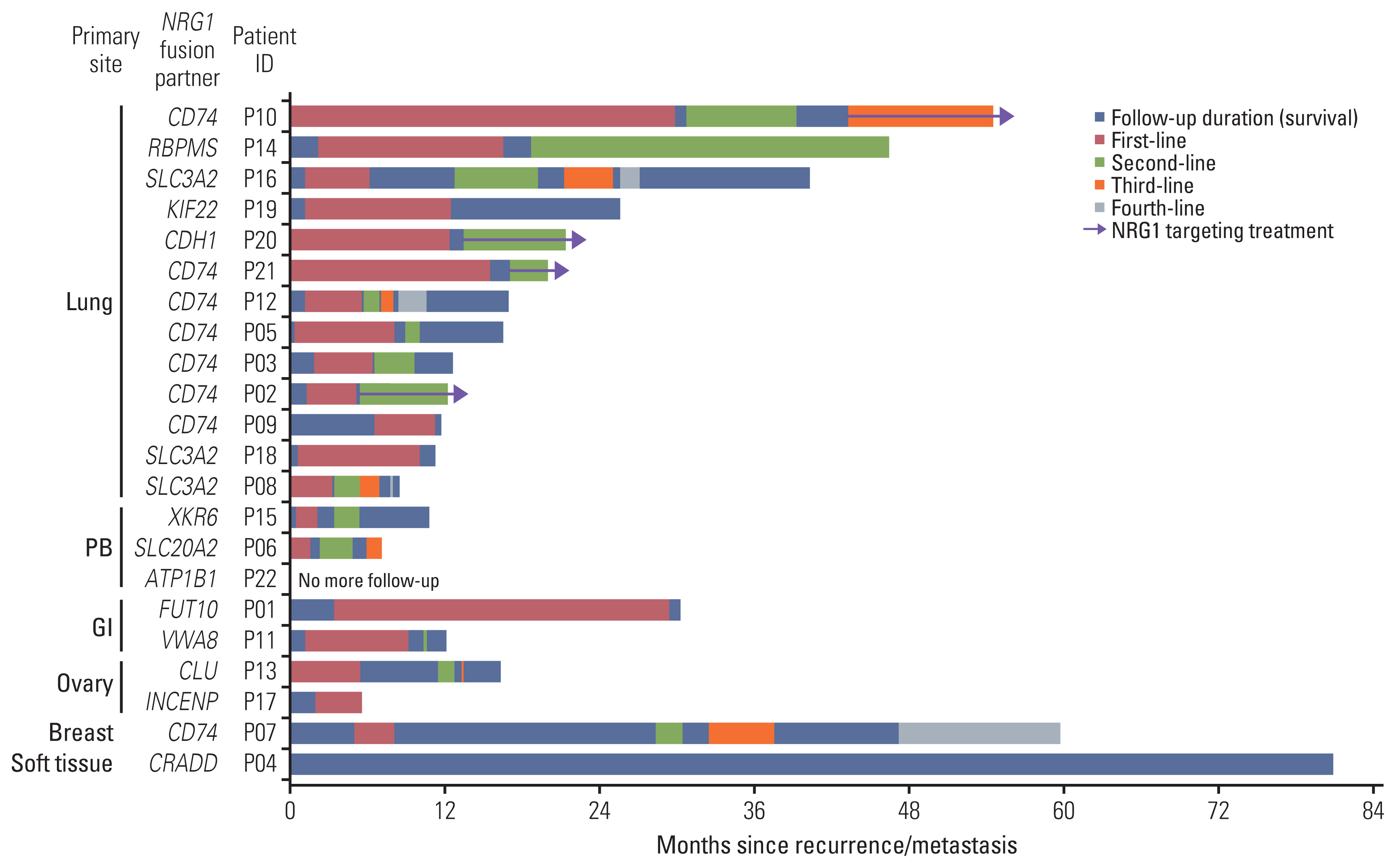
Table 1Characteristics of patients with NRG1 fusion–positive tumors Table 2Pathological features of NRG1 fusion–positive tumors References1. Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45.
2. Schram AM, Chang MT, Jonsson P, Drilon A. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol. 2017;14:735–48.
3. Tzahar E, Levkowitz G, Karunagaran D, Yi L, Peles E, Lavi S, et al. ErbB-3 and ErbB-4 function as the respective low and high affinity receptors of all Neu differentiation factor/heregulin isoforms. J Biol Chem. 1994;269:25226–33.
4. van Lengerich B, Agnew C, Puchner EM, Huang B, Jura N. EGF and NRG induce phosphorylation of HER3/ERBB3 by EGFR using distinct oligomeric mechanisms. Proc Natl Acad Sci U S A. 2017;114:E2836–45.
5. Liu X, Baker E, Eyre HJ, Sutherland GR, Zhou M. Gamma-heregulin: a fusion gene of DOC-4 and neuregulin-1 derived from a chromosome translocation. Oncogene. 1999;18:7110–4.
6. Schaefer G, Fitzpatrick VD, Sliwkowski MX. Gamma-heregulin: a novel heregulin isoform that is an autocrine growth factor for the human breast cancer cell line, MDA-MB-175. Oncogene. 1997;15:1385–94.
7. Huang HE, Chin SF, Ginestier C, Bardou VJ, Adelaide J, Iyer NG, et al. A recurrent chromosome breakpoint in breast cancer at the NRG1/neuregulin 1/heregulin gene. Cancer Res. 2004;64:6840–4.
8. Adelaide J, Huang HE, Murati A, Alsop AE, Orsetti B, Mozziconacci MJ, et al. A recurrent chromosome translocation breakpoint in breast and pancreatic cancer cell lines targets the neuregulin/NRG1 gene. Genes Chromosomes Cancer. 2003;37:333–45.
9. Fernandez-Cuesta L, Plenker D, Osada H, Sun R, Menon R, Leenders F, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov. 2014;4:415–22.
10. Shim HS, Kenudson M, Zheng Z, Liebers M, Cha YJ, Hoang Ho Q, et al. Unique genetic and survival characteristics of invasive mucinous adenocarcinoma of the lung. J Thorac Oncol. 2015;10:1156–62.
11. Drilon A, Duruisseaux M, Han JY, Ito M, Falcon C, Yang SR, et al. Clinicopathologic features and response to therapy of NRG1 fusion-driven lung cancers: the eNRGy1 global multicenter registry. J Clin Oncol. 2021;39:2791–802.
12. Jonna S, Feldman RA, Swensen J, Gatalica Z, Korn WM, Borghaei H, et al. Detection of NRG1 gene fusions in solid tumors. Clin Cancer Res. 2019;25:4966–72.
13. Cadranel J, Liu SV, Duruisseaux M, Branden E, Goto Y, Weinberg BA, et al. Therapeutic potential of afatinib in NRG1 fusion-driven solid tumors: a case series. Oncologist. 2021;26:7–16.
14. Fontana E, Torga G, Fostea R, Cleator S, Wasserman E, Murat A, et al. Sustained tumor regression with zenocutuzumab, a bispecific antibody targeting human epidermal growth factor receptor 2/human epidermal growth factor receptor 3 signaling, in NRG1 fusion-positive, estrogen receptor-positive breast cancer after progression on a cyclin-dependent kinase 4/6 inhibitor. JCO Precis Oncol. 2022;6:e2100446.
15. Schram AM, Odintsov I, Espinosa-Cotton M, Khodos I, Sisso WJ, Mattar MS, et al. Zenocutuzumab, a HER2xHER3 bispecific antibody, is effective therapy for tumors driven by NRG1 gene rearrangements. Cancer Discov. 2022;12:1233–47.
16. Yatabe Y, Mitsudomi T, Takahashi T. TTF-1 expression in pulmonary adenocarcinomas. Am J Surg Pathol. 2002;26:767–73.
17. Matsubara D, Soda M, Yoshimoto T, Amano Y, Sakuma Y, Yamato A, et al. Inactivating mutations and hypermethylation of the NKX2-1/TTF-1 gene in non-terminal respiratory unit-type lung adenocarcinomas. Cancer Sci. 2017;108:1888–96.
18. Park WY, Kim MH, Shin DH, Lee JH, Choi KU, Kim JY, et al. Ciliated adenocarcinomas of the lung: a tumor of non-terminal respiratory unit origin. Mod Pathol. 2012;25:1265–74.
19. Park E, Shim HS. Detection of targetable genetic alterations in Korean lung cancer patients: a comparison study of single-gene assays and targeted next-generation sequencing. Cancer Res Treat. 2020;52:543–51.
20. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23.
21. Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Kallberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–2.
22. Uhrig S, Ellermann J, Walther T, Burkhardt P, Frohlich M, Hutter B, et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res. 2021;31:448–60.
23. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
24. Wen D, Suggs SV, Karunagaran D, Liu N, Cupples RL, Luo Y, et al. Structural and functional aspects of the multiplicity of Neu differentiation factors. Mol Cell Biol. 1994;14:1909–19.
25. Geles A, Gruber-Moesenbacher U, Quehenberger F, Manzl C, Al Effah M, Grygar E, et al. Pulmonary mucinous adenocarcinomas: architectural patterns in correlation with genetic changes, prognosis and survival. Virchows Arch. 2015;467:675–86.
26. Snyder EL, Watanabe H, Magendantz M, Hoersch S, Chen TA, Wang DG, et al. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell. 2013;50:185–99.
27. Heining C, Horak P, Uhrig S, Codo PL, Klink B, Hutter B, et al. NRG1 fusions in KRAS wild-type pancreatic cancer. Cancer Discov. 2018;8:1087–95.
28. Shin DH, Kim SH, Choi M, Bae YK, Han C, Choi BK, et al. Oncogenic KRAS promotes growth of lung cancer cells expressing SLC3A2-NRG1 fusion via ADAM17-mediated shedding of NRG1. Oncogene. 2022;41:280–92.
|
|
|||||||||||||||||||||||||||||||||||||||||