AbstractPurposeWe aimed to evaluate whether the addition of pemetrexed is effective in improving progression-free survival (PFS) in epidermal growth factor receptor (EGFR)–mutated patients with or without concomitant alterations.
Materials and MethodsThis multicenter clinical trial was conducted in China from June 15, 2018, to May 31, 2019. A total of 92 non–small cell lung cancer (NSCLC) patients harboring EGFR-sensitive mutations were included and divided into concomitant and non-concomitant groups. Patients in each group were randomly treated with EGFR–tyrosine kinase inhibitor (TKI) monotherapy or EGFR-TKI combined with pemetrexed in a ratio of 1:1. PFS was recorded as the primary endpoint.
ResultsThe overall median PFS of this cohort was 10.1 months. There were no significant differences in PFS between patients with and without concomitant and between patients received TKI monotherapy and TKI combined with pemetrexed (p=0.210 and p=0.085, respectively). Stratification analysis indicated that patients received TKI monotherapy had a significantly longer PFS in non-concomitant group than that in concomitant group (p=0.002). In concomitant group, patients received TKI combined with pemetrexed had a significantly longer PFS than patients received TKI monotherapy (p=0.013). Molecular dynamic analysis showed rapidly emerging EGFR T790M in patients received TKI monotherapy. EGFR mutation abundance decreased in patients received TKI combined chemotherapy, which supports better efficacy for a TKI combined chemotherapy as compared to TKI monotherapy. A good correlation between therapeutic efficacy and a change in circulating tumor DNA (ctDNA) status was found in 66% of patients, supporting the guiding role of ctDNA minimal residual disease (MRD) in NSCLC treatment.
IntroductionNon–small cell lung cancer (NSCLC) is the most common subtype of lung cancer, and epidermal growth factor receptor (EGFR) is a major oncogene that occurs in approximately 10%–30% of NSCLC patients [1]. Although first-generation EGFR–tyrosine kinase inhibitor (TKI) have become the standard treatment for NSCLC patients harboring a 19_ Del or L858R mutation for EGFR, most patients develop acquired resistance following 12–24 months of treatment [2]. Previous studies have indicated that the mechanisms for EGFR-acquired resistance include an EGFR T790M mutation, a MET amplification, a PTEN deletion, a PIK3CA mutation, an FGFR1 activation mutation, an epithelial-to-mesenchymal transition, and a transformation to small cell lung cancer [2–7]. Duan et al. [8] stratified NSCLC patients into the following three groups: (1) patients with only an EGFR-sensitive mutation, (2) patients with an EGFR-sensitive mutation and a tumor suppressor gene mutation, and (3) patients with an EGFR-sensitive mutation and multiple oncogene mutations. Their results indicated that during a TKI monotherapy treatment, patients with only an EGFR-sensitive mutation had the best treatment outcome, while patients with an EGFR-sensitive mutation and multiple oncogene mutations had the worst treatment outcome [8]. Studies have also revealed that NSCLC patients with co-mutations in EGFR, as well as in genes such as TP53, KRAS, PIK3CA, and MLH1 may enhance tumor progression over the course of an EGFR-TKI treatment [9–12]. In EGFR-sensitive patients, nearly 8.9%, 3.5%, 6%–36%, and 32.4% of them harbored MLH1 V384Dmutation, PIK3CA mutation, KRAS mutation, and TP53 exon 8 mutation, respectively [9,13,14]. All of these studies support the idea that the molecular stratification of NSCLC patients is of great significance for guiding treatment. How to best serve such patients remains controversial. Nevertheless, the development of appropriate treatments for stratified NSCLC patients is urgently needed.
Chemotherapy is widely used in lung cancer patients and is one of the regimens employed when TKI-resistance occurs. The main molecular target of pemetrexed chemotherapy is thymidylate synthase. A previous study indicated that the combination therapy of pemetrexed and erlotinib is capable of reducing the expression of thymidylate synthase in NSCLC [15]. Cell experiments and animal xenografts have confirmed that pemetrexed can effectively prevent the development of an EGFR T790M mutation [16]. Studies have also shown that a TKI combined with chemotherapy results in better clinical benefits for NSCLC patients harboring EGFR mutations, although the survival benefit of this regimen still needs to be elucidated [17].
In this study, we sought to explore the therapeutic effects of EGFR-TKIs combined with chemotherapy in EGFR-mutated, advanced NSCLC patients having multiple mutations. Using next-generation sequencing (NGS), we also sought to identify the subgroup of patients more likely to respond to EGFR-TKIs combined with chemotherapy, with the ultimate goal of providing evidence for personalized advanced NSCLC treatment.
Materials and Methods1. Study designFor our study, we recruited EGFR-mutated, locally advanced, or metastatic NSCLC patients from the First People’s Hospital of Foshan and the Nanhai People’s Hospital, and compared an EGFR-TKI monotherapy treatment versus an EGFR-TKI treatment combined with chemotherapy for patients with and without concomitant alterations. Our study was approved by the institutional ethics committees of the First People’s Hospital of Foshan and the Nanhai People’s Hospital. Our study was also conducted in accordance with the Declaration of Helsinki and international standards for good clinical practice. All patients provided written informed consent before entering our study.
2. Study populationFrom June 15, 2018, to May 31, 2019, a total of 286 advanced lung cancer patients were recruited. Patients with an EGFR-sensitive mutation (an exon 19 deletion or an exon 21 L858R mutation, excluding the T790M mutation) were collected from the First People’s Hospital of Foshan and the Nanhai People’s Hospital. Patients were excluded if they had primary surgery or received prior systemic treatment including chemotherapy, target therapy, or immunotherapy. Patients with ALK, MET, ROS1, and RET position variants were also excluded. Based on these criteria, 92 patients were eligible for subsequent analysis. Patients with MLH1 V384D/TP53 exon 8/KRAS/PIK3CA alterations were classified as the concomitant arm, while those without concomitant alterations were classified as the non-concomitant arm (Fig. 1).
3. Random assignmentPatients from the two study arms were randomly assigned to receive EGFR-TKI alone or EGFR-TKI combined with pemetrexed at a ratio of 1:1, so as to obtain more reliable efficacy and safety research data. Random assignment was conducted using a computer-generated random sequence and an interactive voice-response system and was stratified by sex (male vs. female).
4. Treatment protocolEGFR-TKIs utilized included gefitinib or icotinib. Standard treatment courses and dosages, based on the corresponding drug’s instruction manual, were applied. For an EGFR-TKI combined with pemetrexed treatment, pemetrexed was given on the first day of each cycle (21 days) at a dose of 500 mg/m2. All treatments were continued until disease progression, an unacceptable adverse event occurred, or an interruption due to other factors occurred.
5. Outcome measuresTumor responses at the baseline and during treatment were recorded and evaluated based on Response Evaluation Criteria in Solid Tumor 1.1 criterion. Patient re-examination was performed every six weeks. Therapeutic outcomes were evaluated with imaging of computerized tomography. The primary endpoint for our study was progression-free survival (PFS), which was defined as the time from the point of data for initial treatment until the point of data for progressive disease, as determined using a radiologic evaluation or death from any cause. Adverse events were evaluated according to National Cancer Institute Common Terminology Criteria for Adverse Events (ver. 4.0).
6. Sample collectionFormalin-fixed, paraffin-embedded (FFPE) tumor tissues and matched blood samples were collected and transferred to OrigiMed Co., Ltd. (Shanghai, China) for genomic testing. In addition to the baseline genomic testing of tumor tissues, plasma samples collected at 2, 4, and 8 cycles of treatment were used for circulating tumor DNA (ctDNA) testing. Genomic DNA and ctDNA were prepared using the QIAamp DNA FFPE Tissue Kit and the QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany), respectively, according to the manufacturer’s instructions. The concentration of DNA was measured using Qubit and normalized to 20–50 ng/μL.
7. The identification of genomic alterationsGenomic mutations were identified using the NGS-based YuanSu 450 gene panel (OrigiMed), which covers all of the coding exons for 450 cancer-related genes, as well as 64 selected introns in 39 genes that are frequently rearranged in solid tumors. Genes were captured and sequenced at a mean depth of 800× using an Illumina NextSeq 500 (Illumina, San Diego, CA). Dynamic detection of plasma ctDNA was performed using an NGS-based, XiYuan gene panel (OrigiMed) that included EGFR and 17 additional actionable target genes. Detailed procedures employed for identifying genomic alterations were, as follows: (1) single nucleotide variants (SNVs) were identified using MuTect (v1.17); (2) insertion-deletion (Indels) were identified using PINDEL (v2.05); (3) copy number variation regions were identified using Control-FREEC (v9.7), based on the following parameters: window=50,000 and step=10,000; (4) gene fusions were detected using an in-house pipeline; and (5) gene rearrangements were assessed with the Integrative Genomics Viewer (IGV). The functional impact of each genomic alteration was annotated using SnpEff3.0.
8. Statistical analysisStatistical analyses were performed using SPSS ver. 22.0 (IBM Corp., Armonk, NY). Fisher’s exact test was used for significant difference analyses. A Kaplan-Meier curve and a log-ranked test were used for the survival analysis. p < 0.05 was considered to be statistically significant.
Results1. Patient characterizationA total of 92 NSCLC patients harboring EGFR-sensitive mutations, including 19_Del and L858R, were eligible for subsequent analysis. The cohort consisted of 31 (34%) male and 61 (66%) female patients, with a median age of 65 years old. All patients had advanced NSCLC, including four patients (4%) at stage III and 88 (96%) patients at stage IV, as identified using American Joint Committee on Cancer, 8th edition, criteria. Forty-five patients received an EGFR-TKI (icotinib or gefitinib) monotherapy and 47 patients received an EGFR-TKI treatment combined with pemetrexed (Table 1).
2. Genomic alterations in EGFR-mutated NSCLC patientsAccording to the genomic alterations from the baseline tumor tissue samples, 497 clinically relevant genomic alterations in 205 genes were identified in addition to EGFR alterations. The most commonly mutated genes included TP53 (70%), TERT (15%), CDKN2A (12%), MDM2 (11%), RBM10 (11%), SDHA (11%), and NKX2-1 (10%) (S1 Fig.). The most common types of TP53 mutations were SNVs/Indels and 29% (19/65) of them were TP53 exon 8 mutations. In addition, the mutational frequency of MLH1 V348D and PIK3CA were both 8% (7/92) in this cohort. No concurrent KRAS mutation was detected in this cohort. Nearly 29% (27/92) of patients harbored concomitant alterations with at least one mutation for MLH1 V384D, TP53 exon 8, and PIK3CA.
3. An EGFR-TKI combined with pemetrexed is more suitable for patients with concomitant alterationsIn our cohort, there were 27 patients with concomitant alterations, including 14 patients received TKI monotherapy and 13 patients received TKI combined chemotherapy, and 65 patients without concomitant alterations, including 31 patients received TKI monotherapy, and 34 patients received TKI combined chemotherapy. The overall median PFS of this cohort was 10.1 months (95% confidence interval [CI], 9.2 to 13.2) (S2 Fig.). For patients without concomitant alterations, median PFS was 11.2 months (95% Cl, 9.2 to 14.2), no significant difference was determined for patients treated with an EGFR-TKI monotherapy or for patients with an EGFR-TKI treatment combined with pemetrexed treatment (p=0.910) (Fig. 2A). While for patients with concomitant alterations, the median PFS was 9.7 months (95% Cl, 7.8 to 13.5), and notably, the PFS of patients with EGFR-TKI treatment combined with pemetrexed treatment was significantly longer than those with the TKI monotherapy treatment (p=0.013) (Fig. 2B). Using a stratified analysis, we determined that for patients with an EGFR-TKI monotherapy treatment, PFS was significantly worse in patients with concomitant alterations than patients without concomitant alteration (p=0.002) (Fig. 2C), while in patients with EGFR-TKI combined pemetrexed treatment, no significant PFS difference was observed between patients with and without concomitant alterations (p=0.780) (Fig. 2D). However, we did not find a significant difference in prognosis between the TKI monotherapy group and the TKI combined pemetrexed treatment group (9.8 vs. 10.5 months, p=0.085) (Fig. 2E). The results suggest that concomitant alteration status may impact the efficacy of EGFR-TKI treatment, and support our hypothesis that an EGFR-TKI treatment combined with pemetrexed may effectively improve the prognosis of patients with concomitant alterations.
4. Molecular dynamic analysis reveals the advantages of EGFR-TKI combined with chemotherapy and the guiding role of ctDNA status in efficacy prediction of patientsTo better understand the impact of concomitant alteration status and different treatment regimens on the prognosis of patients, we analyzed the development of EGFR T790M mutations during the course of treatment. By comparing patients with and without concomitant alterations, we found, for all monitoring stages, that the detection rate of T790M mutation in patients with concomitant alterations was lower than that in patients without concomitant alterations. However, no significant difference was determined (Fig. 3A). For the EGFR-TKI monotherapy group, the detection rate of T790M mutation during the first monitoring stage (1–4 months) quickly increased, and gradually decreased during the second (4–8 months) and third (8–12 months) stages. For an EGFR-TKI treatment combined with pemetrexed group, the detection rate of T790M mutation was low and was relatively consistent throughout the three monitoring stages (Fig. 3B).
Based on a dynamic analysis on EGFR mutation abundance, we found that EGFR mutation abundances significantly decreased during the first monitoring stage, both in patients receiving an EGFR-TKI monotherapy or an EGFR-TKI treatment combined with pemetrexed. In patients treated with an EGFR-TKI monotherapy, the EGFR mutation abundance began to increase during the second monitoring stage and reached its highest value during the third monitoring stage. For patients treated with an EGFR-TKI combined with pemetrexed, EGFR mutation abundance remained low during the second monitoring stage and began to increase during the third monitoring stage. Although EGFR abundance increased during the third monitoring stage, the level was still equivalent to that seen during the first monitoring stage (Fig. 3C).
We further evaluated ctDNA status in collected blood samples obtained from 32 enrolled patients by calculating the average variant allele frequency (VAF). Among them, seven patients with concomitant alterations and 11 patients without concomitant alterations displayed disease progress, and three patients with and 11 patients without concomitant alterations displayed disease stability. No significant correlation was determined between treatment strategy and disease status. For patients with disease progress, ctDNA VAF increased during dynamic monitoring in 10 out of 18 patients (Fig. 4). Although the remaining eight patients did not display a ctDNA VAF increase, in six patients, ctDNA VAF was not detected for over half a year until disease progressed. In a similar manner, 11 of 14 patients with disease stability displayed a decline in ctDNA during dynamic monitoring, while the remaining three patients displayed an increase in ctDNA VAF (Fig. 4). These three patients did not have a ctDNA test for more than ten months, from the last detection to the latest follow-up. Taken together, our results indicate that treatment efficiency in nearly 66% (21/32) of patients was consistent with ctDNA status.
5. Adverse eventsIn this cohort, a total of 46 patients experienced adverse events, of which 21 received an EGFR-TKI monotherapy treatment and 25 received an EGFR-TKI treatment combined with pemetrexed. Amongst patients receiving an EGFR-TKI monotherapy, four exhibited high-grade adverse events, including one patient with acne-like dermatitis, one patient with elevated aspartate aminotransferase (AST), one patient with elevated alanine aminotransferase (ALT), and one patient with diarrhea. Amongst 25 patients receiving an EGFR-TKI treatment combined with chemotherapy, eight patients exhibited high-grade adverse events, including two patients with acne-like dermatitis, one patient with elevated AST, one patient with elevated ALT, one patient with stomatitis, one patient with anemia, one patient with diarrhea, and one patient with neutropenia.
DiscussionFirst-generation EGFR-TKIs such as gefitinib, erlotinib, and icotinib have been widely used in clinics for treating advanced NSCLC patients [18]. Acquired resistance to TKIs is one of the main challenges for the clinical management of NSCLC [2]. In our study, 29.3% (27/92) of enrolled patients displayed concomitant alterations with at least one mutation for MLH1 V384D, TP53 exon 8, and PIK3CA. A survival analysis indicated that patients with concomitant alterations had a worse prognosis as compared to patients without concomitant alterations. This result is consistent with previous studies [9,10,12], and suggests that concomitant mutations, including MLH1 V348D, TP53 exon 8, and PIK3CA, may be associated with acquired TKI resistance.
Cisplatin-based chemotherapy is the first-line strategy for treating wild-type EGFR NSCLC [19]. Considering differences in drug mechanisms, EGFR-TKIs combined with chemotherapy for treating advanced lung cancer may result in better efficacy [20,21]. A randomized, Phase II clinical study found that EGFR-TKI combined with pemetrexed significantly prolonged PFS, time to progression, and the duration of response in advanced, non-squamous NSCLC patients with an EGFR activation mutation [17]. Although no significant difference was determined for overall survival, another randomized Phase II clinical trial investigating the first-line treatment of advanced NSCLC found that erlotinib combined with gemcitabine and carboplatin significantly prolonged the PFS of patients with an EGFR activation mutation as compared to the lone use of gemcitabine and carboplatin chemotherapy [22].
Recently, a meta-analysis indicated that first-generation TKIs combined with platinum-based doublet chemotherapeutic drugs are more likely to benefit advanced NSCLC patients with EGFR-sensitive mutations [23]. However, using doublet chemotherapy portends that patients will suffer greater chemotherapy toxicity. Considering the toxic and side effects and drug efficacy, pemetrexed was selected to combine with TKI in the study to compare the prognosis of patients treated with monotherapy and combination therapy. Similar to previous studies [17,24], although no statistically significant difference was determined, the survival curve displayed a slight difference.
Several past studies have yielded no significant difference when employing a TKI combined with chemotherapy and when employing chemotherapy alone in unselected patients [25]. An inadequate selection of patients and an interference in the cell cycle time between a TKI and chemotherapy have been hypothesized to be reasons for these types of negative results [21]. In this cohort, nearly 52% (48/92) of patients carried the EGFR L858R mutation. A previous study has shown that patients harbored EGFR 19_Del confers lower PFS with icotinib than those harbored EGFR L858R mutations [26]. Therefore, the distribution of EGFR L858R and EGFR 19_Del may be the cause of a shorter PFS than previous studies. In addition, previous studies indicate that mutations in TP53 exon 8, PIK3CA, and MLH1 V348D may be associated with the acquired resistance related to EGFR-TKI treatment [9,10,12]. For our study, we not only enrolled advanced NSCLC patients with activating EGFR mutations but also stratified patients based on potential influencing factors such as TP53 exon 8, PIK3CA, and MLH1 V348D. This may be another potential reason why our results are different from those obtained from previous studies. Our results also revealed significantly better prognosis for the TKI combined with chemotherapy group for patients with concomitant alterations, as compared to the TKI monotherapy group, suggesting that the non-specific killing effect of chemotherapy is effective in selected patients with an EGFR-sensitive mutation and concomitant alterations including TP53 exon 8, PIK3CA, or MLH1 V348D, potentially resulting in a benefit from combination therapy for nearly 30% of Asian lung cancer patients. In addition, EGFR T790M accounts for about 50% of the resistance mechanism of first-generation EGFR-TKIs [27]. Although osimertinib shows activity in patients with acquired T790M mutation and is recommended as the first-line treatment against NSCLC with TKI-sensitive EGFR mutations [28], our results also suggest that osimertinib combined with chemotherapy may be a potential strategy for NSCLC patients harboring an EGFR mutation.
In addition to the tendency of a TKI combined with pemetrexed to improve PFS in non-stratified patients and the finding that a TKI combined with pemetrexed significantly improved PFS for patients with concomitant alterations, we also observed similar efficacy between the TKI monotherapy group and the TKI combined with chemotherapy group for patients without concomitant alterations. Without considering concomitant alterations, Cheng et al. [17] reported that, compared to gefitinib monotherapy, the combination of gefitinib and pemetrexed effectively improves PFS in NSCLC patients. Based on previous studies [8,17], we fully considered the effects of different patient stratifications in regards to a TKI monotherapy and a TKI combined with chemotherapy, and deduced that whether or not an EGFR-sensitive patient has concomitant alterations plays an important guiding role in relation to further combined TKI and chemotherapy treatments. However, having a small number of patients was a limitation of our study. Further clinical trials with an enlarged cohort are required.
EGFR-activated tumors are composed of heterogeneous subclones containing a variety of EGFR allele combinations. Changes in ctDNA levels are known to be correlated with clinical responses to an EGFR-TKI. Dynamic monitoring of ctDNA levels can provide real-time tumor responses to an EGFR-TKI and can lead to information regarding the proportion of each subclone [29]. Past studies have also indicated that almost half of patients with an EGFR T790M mutation display clinical progression around 2.2 months [30]. Based on ctDNA detection, a previous study showed that the PFS of patients with a high EGFR mutation abundance was better than that of patients with a low EGFR mutation abundance, suggesting that the dynamic quantitative analysis of an EGFR mutation in ctDNA can be used to guide for the personalized treatment of advanced lung adenocarcinoma [31].
In our study, dynamic monitoring results revealed that the EGFR T790M mutation appeared earlier within the TKI monotherapy group, indicating a worse prognosis for TKI monotherapy as compared to a TKI combined with chemotherapy. Such results are very consistent with our survival analysis. However, the frequency of the T790M mutation was lower in patients with concomitant alterations, a contradiction in regard to the poor prognosis experienced by these patients. Since TP53 exon 8, MLH1 V348D, and PIK3CA serve as potential mechanisms for EGFR-TKI resistance [9,10,12], the acquired EGFR-TKI resistance mechanism, with the exception of T790M, is more likely in patients with concomitant alterations. Taken together, our results indicate the importance of EGFR-sensitive patient stratification and support the notion that the dynamic monitoring of EGFR T790M mutation should be performed with caution when used to guide for personalizing the treatment of patients with concomitant alterations.
Recently, ctDNA minimal residual disease (MRD) has been reported to be a biomarker for early predictions of recurrence in surgically resected NSCLC patients [32]. CtDNA status may also be used to represent MRD levels [32]. However, few reports exist regarding how to guide medication in patients with advanced NSCLC. In this study, we sought to evaluate the therapeutic effect of advanced NSCLC patients by analyzing ctDNA status. Although no correlation between ctDNA status and treatment strategy existed for ctDNA and concomitant alterations status, in 66% of our advanced NSCLC patients, we did find that changes in ctDNA status were associated with treatment responses. For most of the remaining patients, a ctDNA determination was not quickly obtained subsequent to the previous ctDNA determination and the last follow-up interval, which may be a reason for consistency in regards to ctDNA status and therapeutic effects. Furthermore, a previous study speculated that MRD caused by intratumoral heterogeneity may be the earliest form of acquired drug resistance [33]. Therefore, without personalized ctDNA, the lack of an MRD analysis may be a deficiency of our study and is another potential reason for inconsistencies associated with therapeutic efficacy and ctDNA status. However, the application of ctDNA MRD in the evaluation of therapeutic efficacy for advanced NSCLC is still controversial and requires additional confirmation.
In conclusion, our results suggest that patients with concomitant alterations are more sensitive to an EGFR-TKI treatment combined with chemotherapy, both in terms of prognosis and dynamic monitoring results. Our results also indicate potential evidence for predicting therapeutic efficacy using ctDNA MRD in patients with advanced NSCLC, which provides valuable information for lung cancer precision medicine.
Electronic Supplementary MaterialSupplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
NotesEthical Statement The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethical committees of the First People’s Hospital of Foshan and Nanhai People’s Hospital, and informed consent was obtained from all patients. Author Contributions Conceived and designed the analysis: You D, Gu W (Weiquan Gu), Feng W. Collected the data: Gu W (Weiguang Gu), Lu Y, Li M, Yang S, Liang J, Ye Z, Feng W. Contributed data or analysis tools: Gu W (Weiguang Gu), Zhang H, Lu Y, Li M, Yang S, Liang J, Ye Z, Li Z, He M, Gu W (Weiquan Gu), Feng W. Performed the analysis: Gu W (Weiquang Gu), Zhang H, Shi X, Wang F. Wrote the paper: Gu W (Weiguang Gu), Zhang H, Lu Y, Li M, Yang S, Liang J, Ye Z, Li Z, He M, Shi X, Wang F, You D, Gu W (Weiquan Gu), Feng W. AcknowledgmentsThe authors are grateful to all study participants, patients, and their family members for their contributions and support.
Fig. 1The randomization and trial profile. EGFR, epidermal growth factor receptor; NSCLC, non–small cell lung cancer; TKI, tyrosine kinase inhibitor. 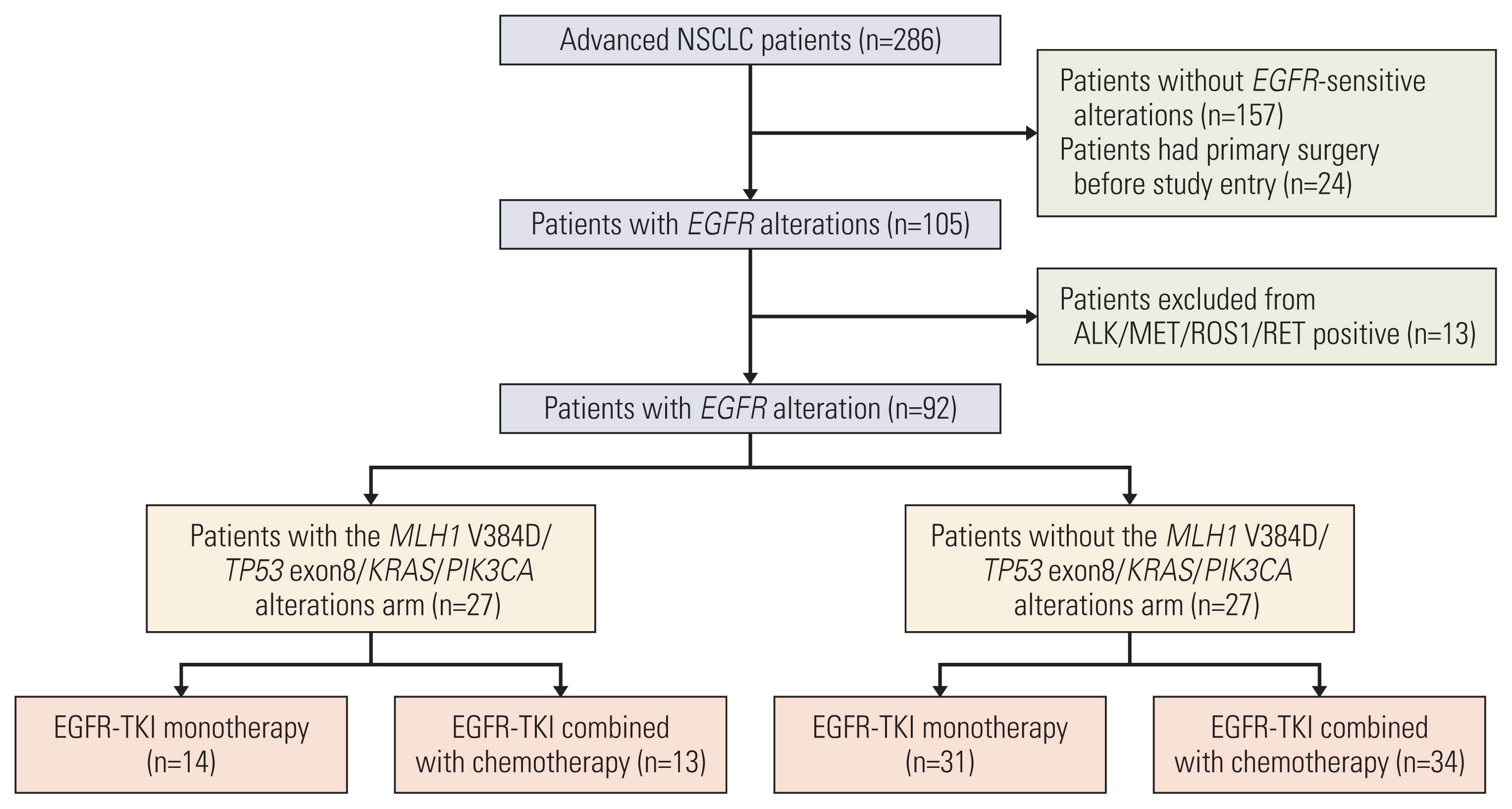
Fig. 2Kaplan-Meier curves for progression free survival (PFS) in patients. (A) Comparison analysis of PFS treated with epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI) and EGFR-TKI combined with pemetrexed in patients without concomitant alterations. (B) Comparison analysis of PFS treated with EGFR-TKI and EGFR-TKI combined with pemetrexed in patients with concomitant alterations. (C) A comparison analysis of PFS between EGFR-TKI treated patients with and without concomitant alterations. (D) A comparison analysis of PFS between EGFR-TKI combined with pemetrexed treated patients with and without concomitant alterations. CI, confidence interval. (E) A comparison analysis of PFS between patients with EGFR-TKI and EGFR-TKI combined with pemetrexed treatments. “Yes” indicates patients with concomitant alterations. “No” indicates patients without concomitant alterations. “TKI” indicates patients that received an EGFR-TKI treatment. “TKI+chemo” indicates patients that received an EGFR-TKI combined with pemetrexed treatment. 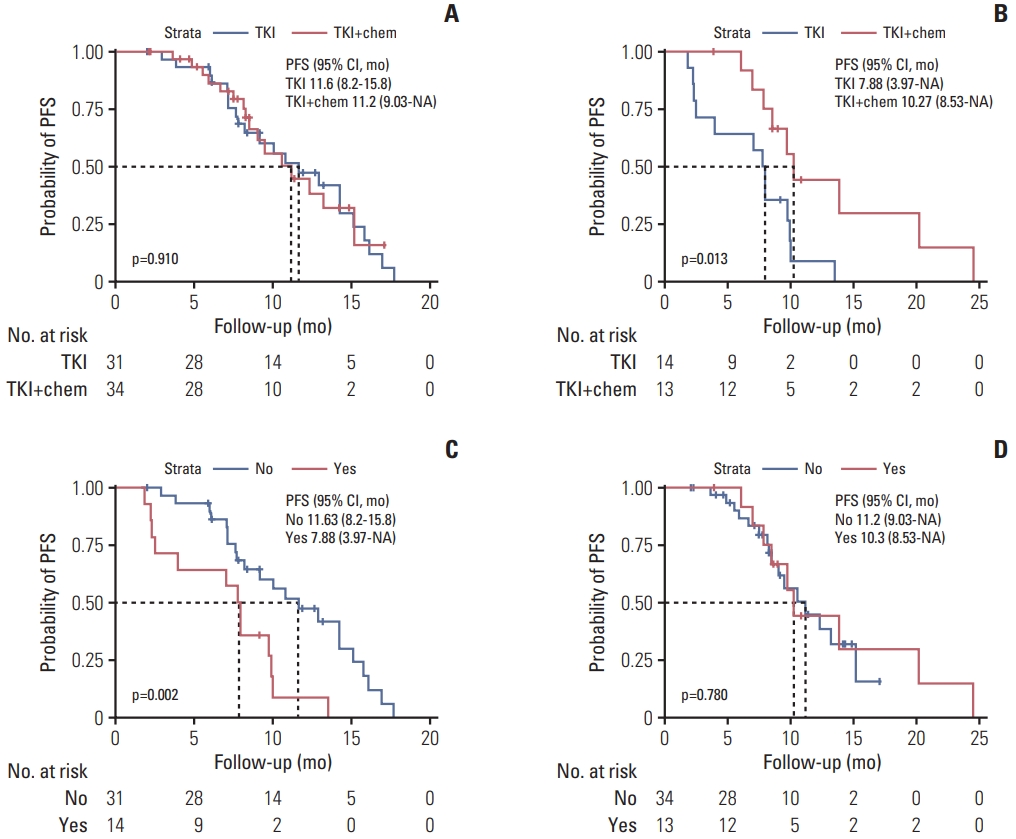
Fig. 3Dynamic circulating tumor DNA (ctDNA) monitoring for epidermal growth factor receptor (EGFR) mutations. (A) The detection rate of EGFR T790M mutation in patients with and without concomitant alterations. (B) The detection rate of EGFR T790M mutation in patients with an EGFR–tyrosine kinase inhibitor (TKI) monotherapy treatment and with EGFR-TKI combined with pemetrexed treatment. (C) Dynamic ctDNA monitoring for EGFR mutation abundance in patients with an EGFR-TKI monotherapy treatment and with EGFR-TKI combined with pemetrexed treatment. “Yes” indicates patients with concomitant alterations. “No” indicates patients without concomitant alterations. “TKI” indicates that patients received an EGFR-TKI treatment. “TKI+chemo” indicates that patients recei-ved an EGFR-TKI combined with pemetrexed treatment. 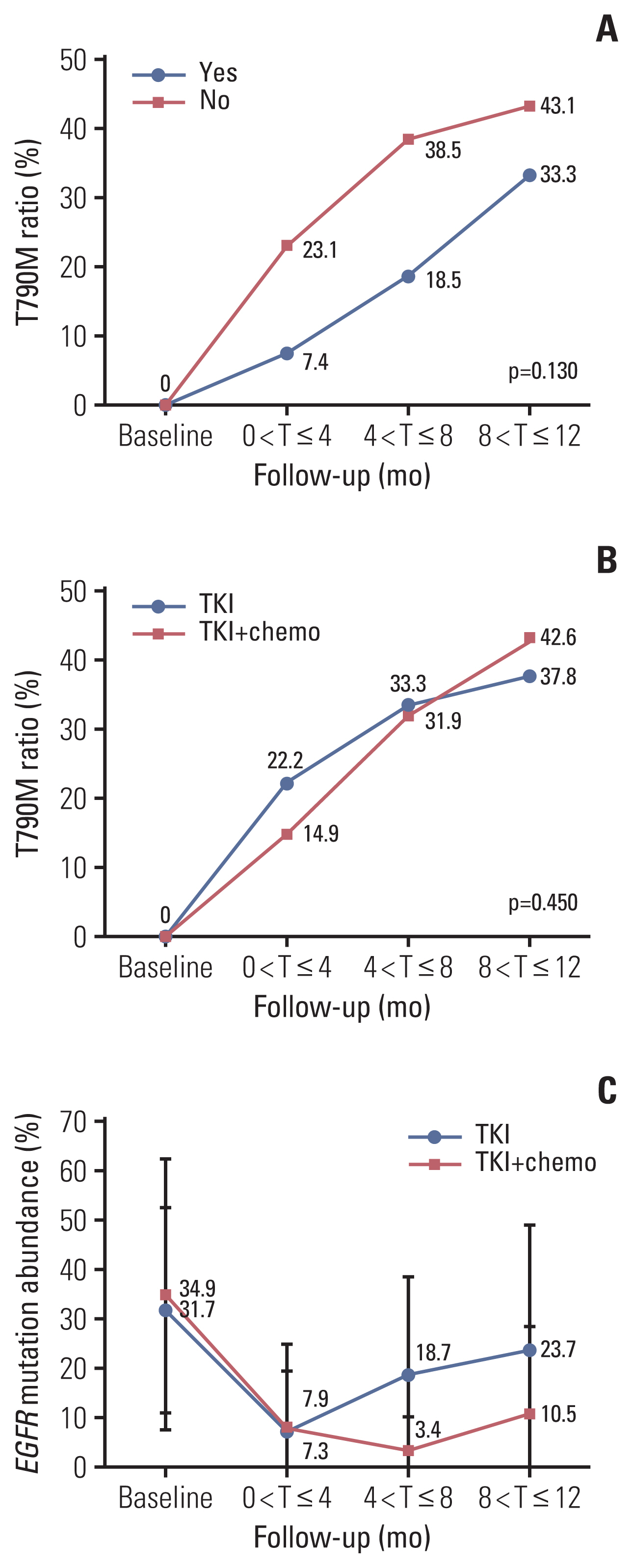
Fig. 4Dynamic circulating tumor DNA (ctDNA) status and clinical outcomes. The x-axis represents the follow-up time (months). The y-axis represents each patient. Circles represent the time point of ctDNA detection. Sizes for individual circles represent different values of ctDNA variant allele frequency (VAF). Red crosses indicate the progress of disease. Green arrows indicate the stability of disease. 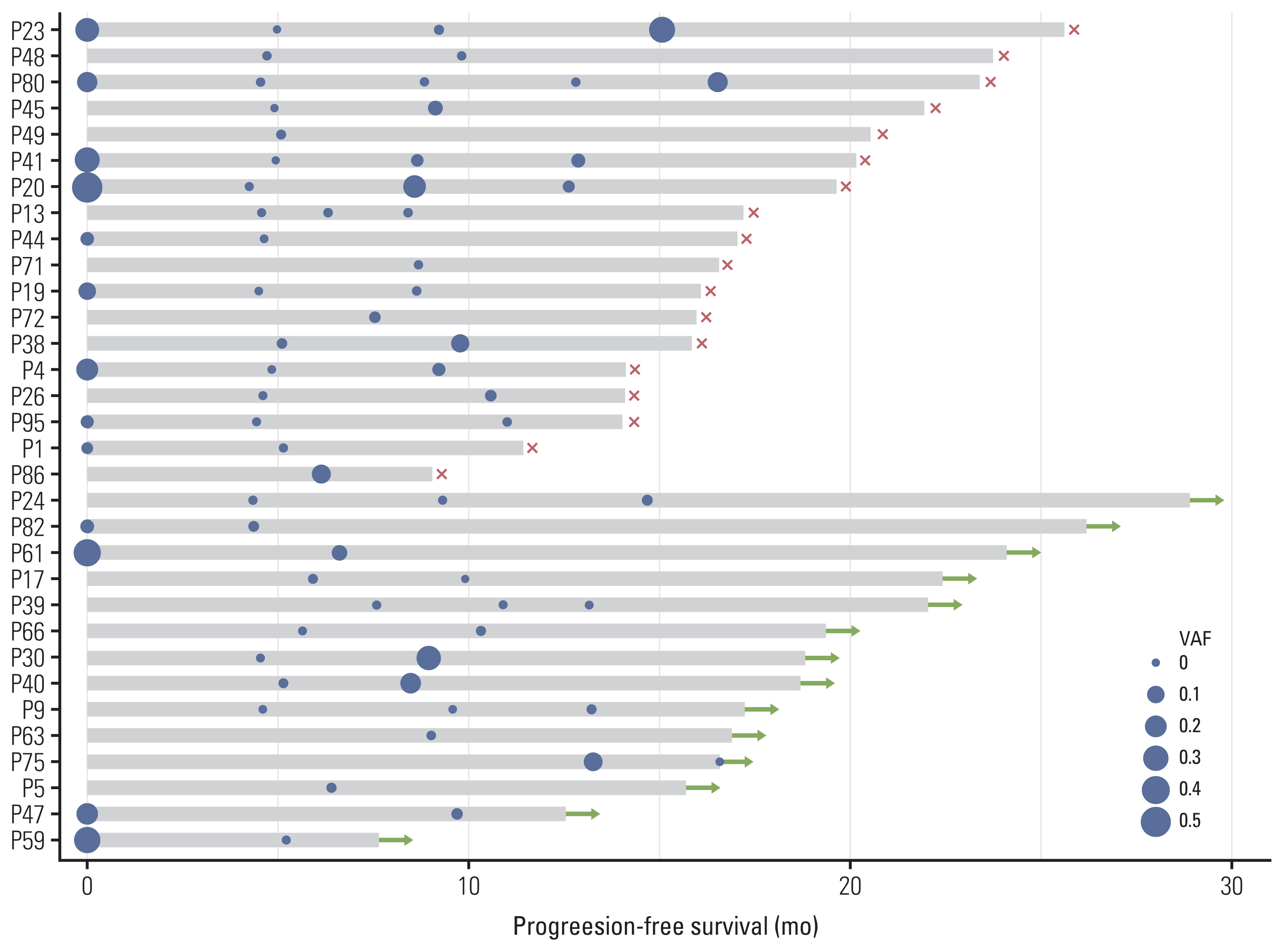
Table 1Characterization of the enrolled patients (n=92) References1. Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81.
2. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–46.
3. Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92.
4. Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43.
5. Wu SG, Chang YL, Yu CJ, Yang PC, Shih JY. The role of PIK3CA mutations among lung adenocarcinoma ptients with primary and acquired resistance to EGFR tyrosine kinase inhibition. Sci Rep. 2016;6:35249.
6. Gammelgaard KR, Vad-Nielsen J, Clement MS, Weiss S, Daugaard TF, Dagnaes-Hansen F, et al. Up-regulated FGFR1 expression as a mediator of intrinsic TKI resistance in EGFR-mutated NSCLC. Transl Oncol. 2019;12:432–40.
7. Mao C, Liao RY, Chen Q. Loss of PTEN expression predicts resistance to EGFR-targeted monoclonal antibodies in patients with metastatic colorectal cancer. Br J Cancer. 2010;102:940.
8. Duan J, Xu J, Wang Z, Bai H, Cheng Y, An T, et al. Refined stratification based on baseline concomitant mutations and longitudinal circulating tumor DNA monitoring in advanced EGFR-mutant lung adenocarcinoma under gefitinib treatment. J Thorac Oncol. 2020;15:1857–70.
9. Chiu CH, Ho HL, Doong H, Yeh YC, Chen MY, Chou TY, et al. MLH1 V384D polymorphism associates with poor response to EGFR tyrosine kinase inhibitors in patients with EGFR L858R-positive lung adenocarcinoma. Oncotarget. 2015;6:8407–17.
10. Canale M, Petracci E, Delmonte A, Bronte G, Chiadini E, Ludovini V, et al. Concomitant TP53 mutation confers worse prognosis in EGFR-mutated non-small cell lung cancer patients treated with TKIs. J Clin Med. 2020;9:1047.
11. Nardo G, Carlet J, Marra L, Bonanno L, Boscolo A, Dal Maso A, et al. Detection of low-frequency KRAS mutations in cfDNA from EGFR-mutated NSCLC patients after first-line EGFR tyrosine kinase inhibitors. Front Oncol. 2020;10:607840.
12. Deng LL, Gao G, Deng HB, Wang F, Wang ZH, Yang Y. Co-occurring genetic alterations predict distant metastasis and poor efficacy of first-line EGFR-TKIs in EGFR-mutant NSCLC. J Cancer Res Clin Oncol. 2019;145:2613–24.
13. Guo Y, Song J, Wang Y, Huang L, Sun L, Zhao J, et al. Concurrent genetic alterations and other biomarkers predict treatment efficacy of EGFR-TKIs in EGFR-mutant non-small cell lung cancer: a review. Front Oncol. 2020;10:610923.
14. Canale M, Petracci E, Delmonte A, Chiadini E, Dazzi C, Papi M, et al. Impact of TP53 mutations on outcome in EGFR-mutated patients treated with first-line tyrosine kinase inhibitors. Clin Cancer Res. 2017;23:2195–202.
15. Scagliotti GV, Ceppi P, Capelletto E, Novello S. Updated clinical information on multitargeted antifolates in lung cancer. Clin Lung Cancer. 2009;10(Suppl 1):S35–40.
16. La Monica S, Madeddu D, Tiseo M, Vivo V, Galetti M, Cretella D, et al. Combination of gefitinib and pemetrexed prevents the acquisition of TKI resistance in NSCLC cell lines carrying EGFR-activating mutation. J Thorac Oncol. 2016;11:1051–63.
17. Cheng Y, Murakami H, Yang PC, He J, Nakagawa K, Kang JH, et al. Randomized phase II trial of gefitinib with and without pemetrexed as first-line therapy in patients with advanced nonsquamous non-small-cell lung cancer with activating epidermal growth factor receptor mutations. J Clin Oncol. 2016;34:3258–66.
18. Lu N, Pan Y, Han X. A retrospective study on the efficacy and prognostic factors of three EGFR-TKI drugs. Mod Oncol. 2018;26:2362–7.
19. Griesinger F, Korol EE, Kayaniyil S, Varol N, Ebner T, Goring SM. Efficacy and safety of first-line carboplatin-versus cisplatin-based chemotherapy for non-small cell lung cancer: a meta-analysis. Lung Cancer. 2019;135:196–204.
20. Li C, Zhang B, Guo J, Hu F, Nie W, Zheng X, et al. Epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs) combined with chemotherapy delay brain metastasis in patients with EGFR-mutant lung adenocarcinoma. Target Oncol. 2019;14:423–31.
21. Rebuzzi SE, Alfieri R, La Monica S, Minari R, Petronini PG, Tiseo M. Combination of EGFR-TKIs and chemotherapy in advanced EGFR mutated NSCLC: review of the literature and future perspectives. Crit Rev Oncol Hematol. 2020;146:102820.
22. Mok TS, Wu YL, Yu CJ, Zhou C, Chen YM, Zhang L, et al. Randomized, placebo-controlled, phase II study of sequential erlotinib and chemotherapy as first-line treatment for advanced non-small-cell lung cancer. J Clin Oncol. 2009;27:5080–7.
23. Wu Q, Luo W, Li W, Wang T, Huang L, Xu F. First-generation EGFR-TKI plus chemotherapy versus EGFR-TKI alone as first-line treatment in advanced NSCLC with EGFR activating mutation: a systematic review and meta-analysis of randomized controlled trials. Front Oncol. 2021;11:598265.
24. Liu S, He Y, Jiang T, Ren S, Zhou F, Zhao C, et al. EGFR-TKIs plus chemotherapy demonstrated superior efficacy than EGFR-TKIs alone as first-line setting in advanced NSCLC patients with EGFR mutation and BIM deletion polymorphism. Lung Cancer. 2018;120:82–7.
25. Gatzemeier U, Pluzanska A, Szczesna A, Kaukel E, Roubec J, De Rosa F, et al. Phase III study of erlotinib in combination with cisplatin and gemcitabine in advanced non-small-cell lung cancer: the Tarceva Lung Cancer Investigation Trial. J Clin Oncol. 2007;25:1545–52.
26. Wang Y, Yuan X, Yang M, Shen Z, Chen H, He X, et al. Efficacy of icotinib, an EGFR tyrosine kinase inhibitor in non-small cell lung cancer patients with exon 19 deletion and exon 21 L858R: a retrospective analysis in China. Pharmacology. 2021;106:658–66.
27. Huang L, Fu L. Mechanisms of resistance to EGFR tyrosine kinase inhibitors. Acta Pharm Sin B. 2015;5:390–401.
28. Ettinger DS, Wood DE, Aggarwal C, Aisner DL, Akerley W, Bauman JR, et al. NCCN guidelines insights: non-small cell lung cancer, version 1.2020. J Natl Compr Canc Netw. 2019;17:1464–72.
29. Vendrell JA, Mau-Them FT, Beganton B, Godreuil S, Coopman P, Solassol J. Circulating cell free tumor DNA detection as a routine tool for lung cancer patient management. Int J Mol Sci. 2017;18:264.
30. Ni J, Weng L, Liu Y, Sun Z, Bai C, Wang Y. Dynamic monitoring of EGFR mutations in circulating cell-free DNA for EGFR-mutant metastatic patients with lung cancer: early detection of drug resistance and prognostic significance. Oncol Lett. 2017;13:4549–57.
31. Li X, Cai W, Yang G, Su C, Ren S, Zhao C, et al. Comprehensive analysis of EGFR-mutant abundance and its effect on efficacy of EGFR TKIs in advanced NSCLC with EGFR mutations. J Thorac Oncol. 2017;12:1388–97.
|
|
|||||||||||||||||||||||||||||||||||||||||