Analysis of Plasma Circulating Tumor DNA in Borderline Resectable Pancreatic Cancer Treated with Neoadjuvant Modified FOLFIRINOX: Clinical Relevance of DNA Damage Repair Gene Alteration Detection
Article information
Abstract
Purpose
There are no reliable biomarkers to guide treatment for patients with borderline resectable pancreatic cancer (BRPC) in the neoadjuvant setting. We used plasma circulating tumor DNA (ctDNA) sequencing to search biomarkers for patients with BRPC receiving neoadjuvant mFOLFIRINOX in our phase 2 clinical trial (NCT02749136).
Materials and Methods
Among the 44 patients enrolled in the trial, patients with plasma ctDNA sequencing at baseline or post-operation were included in this analysis. Plasma cell-free DNA isolation and sequencing were performed using the Guardant 360 assay. Detection of genomic alterations, including DNA damage repair (DDR) genes, were examined for correlations with survival.
Results
Among the 44 patients, 28 patients had ctDNA sequencing data qualified for the analysis and were included in this study. Among the 25 patients with baseline plasma ctDNA data, 10 patients (40%) had alterations of DDR genes detected at baseline, including ATM, BRCA1, BRCA2 and MLH1, and showed significantly better progression-free survival than those without such DDR gene alterations detected (median, 26.6 vs. 13.5 months; log-rank p=0.004). Patients with somatic KRAS mutations detected at baseline (n=6) had significantly worse overall survival (median, 8.5 months vs. not applicable; log-rank p=0.003) than those without. Among 13 patients with post-operative plasma ctDNA data, eight patients (61.5%) had detectable somatic alterations.
Conclusion
Detection of DDR gene mutations from plasma ctDNA at baseline was associated with better survival outcomes of patients with borderline resectable pancreatic ductal adenocarcinoma treated with neoadjuvant mFOLFIRINOX and may be a prognostic biomarker.
Introduction
Pancreatic cancer is a highly aggressive malignancy arising from the exocrine pancreas, with a 5-year survival rate from the time of diagnosis of only 10% [1]. As surgery is the only curative option, pancreatic cancer is classified according to its resectability [2]. The resectability criteria suggested by the National Comprehensive Cancer Network (NCCN) are widely accepted, and only a few patients are diagnosed with initially resectable disease and may undergo upfront surgery [3,4]. For patients with borderline resectable pancreatic cancer (BRPC), neoadjuvant therapy followed by surgical resection is the standard treatment [4].
Modified fluorouracil plus leucovorin, oxaliplatin, and irinotecan (mFOLFIRINOX) is one of the recommended regimens for neoadjuvant treatment of BRPC. In our previous phase 2 clinical trial of 44 patients with BRPC treated with neoadjuvant mFOLFIRINOX followed by surgery and adjuvant gemcitabine, the 1-year progression-free survival (PFS) rate was 52.3% (95% confidence interval [CI], 37.6 to 67.0) and 27 patients (61.4%) received surgical resection [5]. Other treatment regimens, including gemcitabine plus nab-paclitaxel or the addition of subsequent chemoradiation can be used, although there is currently no evidence to guide the selection of specific regimens [6].
Plasma circulating tumor DNA (ctDNA) analysis is widely investigated as a source of biomarkers to guide treatment and predict outcomes of patients with different types of solid tumor [7]. Genomic profiling of cancer based on plasma ctDNA has several advantages compared to tissue-based sequencing, including that plasma ctDNA may reflect the tumor heterogeneity, and it can also be applied in cases when an adequate tissue sample is unavailable [8]. Also, plasma ctDNA analysis may be used to evaluate the minimal residual disease among patients who received curative surgery [9].
Previous studies were performed with plasma ctDNA analysis to evaluate clinical outcomes of neoadjuvant chemotherapy in several types of cancers [10–12]. However, there is only limited evidence on biomarkers associated with the outcomes of patients with BRPC treated with neoadjuvant therapy, and additional plasma ctDNA analysis may provide potential biomarkers to guide the management of BRPC patients. In this study, we conducted plasma ctDNA analysis to search for potential biomarkers for BRPC patients treated with neoadjuvant mFOLFIRINOX in our phase 2 clinical trial (ClinicalTrials.gov identifier: NCT02749136). Alterations of genes involved in the DNA damage repair (DDR) pathway and response to neoadjuvant mFOLFIRINOX were mainly investigated as several previous studies have shown platinum-sensitivity of pancreatic cancer with germline alterations in BRCA1, BRCA2, and PALB2 [13,14].
Materials and Methods
1. Study design and patients
Patients with a histological or cytological diagnosis of pancreatic adenocarcinoma defined as BRPC as per the NCCN resectability criteria were enrolled and treated with neoadjuvant mFOLFIRINOX followed by surgery and adjuvant gemcitabine in a previous single arm, single center phase 2 trial (NCT02749136) [5]. Patients were treated with neoadjuvant mFOLFIRINOX, including 2,400 mg/m2 of fluorouracil, 400 mg/m2 of leucovorin, 85 mg/m2 of oxaliplatin, and 150 mg/m2 of irinotecan on day 1 every 2 weeks for 8 cycles. Surgery followed by adjuvant gemcitabine of 1,000 mg/m2 for three consecutive weeks every 4 weeks up to 6 cycles were given to patients who achieved resectability following neoadjuvant therapy. Details of the trial design and procedures are described in a previous report [5]. Among these patients, those who had plasma ctDNA targeted next-generation sequencing (NGS) with the Guardant 360 platform were included in this study.
2. Plasma ctDNA analysis
Peripheral blood samples were collected at baseline and at the time of first follow-up after surgical resection. All plasma cell-free DNA isolation and sequencing were performed at Guardant Health Inc. and analyzed using the Guardant 360 assay (Redwood City, CA). After the isolation of plasma from the blood sample using centrifugation, cell-free DNA was extracted from the plasma, and 5 to 30 ng of the cell-free DNA was used to generate sequencing libraries enriched by hybridization capture. The libraries were sequenced using the Illumina NextSeq 550 platform (San Diego, CA), and the sequenced data were analyzed using the standard analysis pipeline of the Guardant 360 assay to detect genomic alterations of 74 genes, including ATM, BRCA1, BRCA2, and MLH1 which are well known as genes involved in DDR pathway [15]. Germline or somatic alterations were also defined according to the standard analysis pipeline of the Guardant 360 assay.
3. Clinical outcomes
Associations of clinical outcomes with detection of genomic alterations involved in the DDR pathway and other commonly altered genes in pancreatic cancer, including KRAS and TP53, were analyzed [16]. PFS was defined as the time from the initiation of neoadjuvant therapy to objective disease progression or any cause of death, and overall survival (OS) was defined as the time from the initiation of neoadjuvant treatment to death from any cause. The best objective response was assessed according to the Response Evaluation Criteria in Solid Tumors (RECIST) ver. 1.1. For patients who received surgical resection, disease-free survival (DFS), defined as the time to surgical confirmation of recurrence, was correlated with the detection of somatic alterations in post-surgery samples.
4. Statistical analysis
Genomic profiling was performed with descriptive methods and visualized using the cBio Cancer Genomics Portal [17,18]. Survival curves were estimated with Kaplan-Meier methods and compared using log-rank tests. All reported p-values are two-sided, and p-values less than 0.05 were considered statistically significant. All statistical analyses were performed using R software, ver. 4.2.0 (http://cran.r-project.org/).
Results
1. Patient characteristics
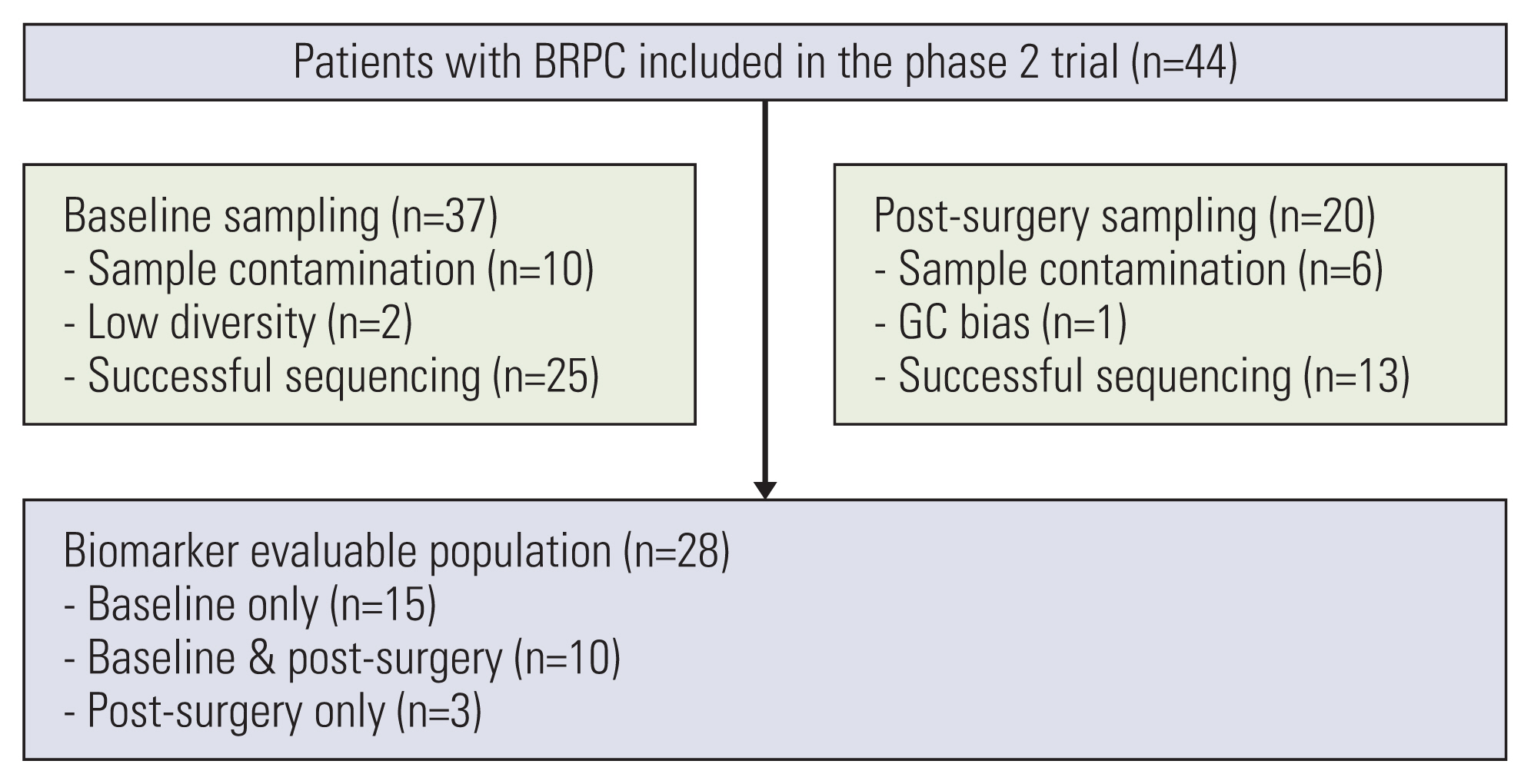
Among the 44 patients with BRPC enrolled in the phase 2 neoadjuvant mFOLFIRINOX trial, 28 patients had plasma ctDNA sequencing data from baseline and/or post-surgery samples available for the analysis and were included in this study (Fig. 1). The availability of baseline and post-surgery data of patients treated in the phase 2 trial is summarized in S1 Table. Of the 28 patients, 25 patients were included in the baseline genomic profiling analysis and 13 patients were included in the post-surgery ctDNA analysis. The baseline patient characteristics are summarized in Table 1. Their median age was 60.5 years (range, 35 to 73 years), and 15 patients (53.6%) were male. All patients had an Eastern Cooperative Oncology Group performance status of 1, and 22 patients (78.6%) had their tumor located in the head of the pancreas. Eleven patients (39.3%) showed a partial response as their best response to neoadjuvant mFOLFIRINOX as per RECIST v1.1. Nineteen patients (67.9%) received surgical resection with curative intent, and R0 resection was achieved in 17 patients (89.5%). All patients received adjuvant gemcitabine after surgery. With a median follow-up duration of 29.0 months (95% CI, 24.6 to not available [NA]), the median PFS of all patients was 14.5 months (95% CI, 13.0 to 26.6).
2. Genomic landscape from baseline plasma ctDNA analysis
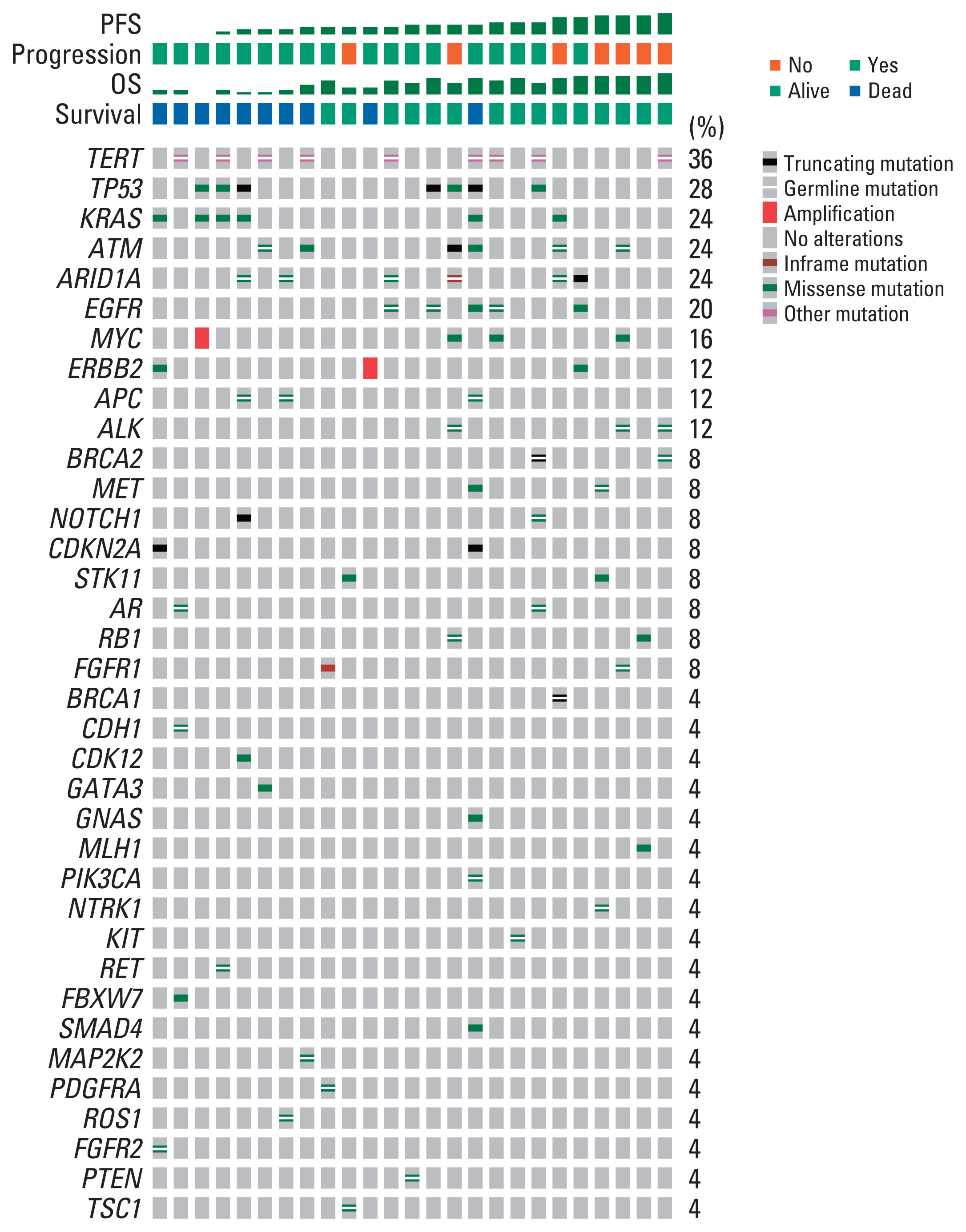
The genomic landscape of the germline and somatic baseline alterations detected by ctDNA analysis is shown in Fig. 2. Among 25 patients included in the baseline analysis, somatic alterations were detected in 14 patients (56.0%) with a median two mutations per patient (range, 1 to 9). The most commonly observed baseline somatic alteration was TP53 (n=7, 28%), followed by KRAS (n=6, 24%). There was no difference in survival outcomes according to detection of somatic alteration at baseline in terms of both PFS (log-rank, p=0.45) and OS (log-rank, p=0.34) (S2 Fig.).
3. Clinical outcomes according to detection of DDR pathway gene alterations
Among 25 patients with baseline plasma ctDNA data, 10 patients (40%) had somatic or germline alterations of DDR pathway genes detected, including ATM (n=7), BRCA1 (n=1), BRCA2 (n=2), and MLH1 (n=1) (S3 Table). Patients with DDR gene alteration detected (n=10) showed significantly longer PFS than those without such DDR gene alterations detected (n=15), with a median PFS of 26.6 months (95% CI, 16.0 to NA) and 13.1 months (95% CI, 7.36 to 18.1), respectively (log-rank, p=0.009) (Fig. 3A). There was a trend toward better survival in terms of OS among patients with DDR pathway gene alterations detected compared to those without (log-rank, p=0.24) (Fig. 3B). When comparing the patient characteristics stratified by detection of the DDR pathway gene alterations, there was no statistically significant difference between the two groups, although patients with DDR pathway gene alterations detected showed a numerically higher objective response rate (40.0% vs. 33.4%) and a higher proportion of patients receiving curative surgery (80.0% vs. 53.3%) (S4 Table).

Kaplan-Meier estimates of survival outcomes according to baseline genomic alterations detected from plasma circulating tumor DNA. Progression-free survival (A) and overall survival (B) according to DNA damage repair (DDR) gene alteration detection. Progression-free survival (C) and overall survival (D) according to KRAS alteration detection. Progression-free survival (E) and overall survival (F) according to TP53 alteration detection.
4. Clinical outcomes according to detection of KRAS and TP53 alterations
Patients with KRAS alterations detected (n=6) tended to have shorter PFS compared to those without (n=19), with a median PFS of 6.8 months (95% CI, 2.8 to NA) and 17.1 months (95% CI, 13.0 to NA), respectively (log-rank, p=0.10) (Fig. 3C). In terms of OS, patients with somatic alterations of the KRAS gene detected at baseline showed significantly worse survival than those without, with a median OS of 8.5 months (95% CI, 7.4 to NA) for patients with KRAS alterations detected, while the median OS was not reached for those without (log-rank, p=0.003) (Fig. 3D). When comparing clinical outcomes according to the detection of somatic TP53 alterations (TP53 alteration not detected [n=18] vs. TP53 alteration detected [n=7]), there was no difference in clinical outcomes in terms of either PFS (log-rank, p=0.26) and OS (log-rank, p=0.25) (Fig. 3E and F).
5. Post-surgery plasma ctDNA analysis and clinical outcomes
Among 13 patients included in the post-surgery analysis, eight patients (61.5%) had somatic alteration detected from plasma following surgery. When comparing DFS from surgical resection according to the detection of somatic alterations in post-surgery samples (detected vs. not detected), there was no significant difference in survival outcomes (log-rank, p=0.39) (S5 Fig.).
Discussion
This retrospective study aimed to assess the utility of plasma ctDNA analysis for patients with BRPC treated with neoadjuvant mFOLFIRINOX. The genetic profiles of the patients from our phase II trial (ClinicalTrials.gov identifier: NCT02749136) were identified using ctDNA analysis with the Guardant 360 assay. Mainly, we analyzed the association of certain mutations, including DDR gene mutations, with OS and PFS to look for potential prognostic biomarkers. We found that the detection of DDR gene (BRCA1, BRCA2, ATM, MLH1) mutations in plasma ctDNA at baseline was significantly associated with better PFS and showed a trend toward better OS. Also, the detection of KRAS mutations in plasma ctDNA at baseline was significantly associated with worse OS and showed a trend toward worse PFS. Importantly, our results highlight the potential utility of pre-treatment ctDNA analysis for patients with BRPC in aiding clinical decisions in the neoadjuvant setting.
However, we did not observe a significant association of post-operative detection of somatic alterations on ctDNA analysis with survival. Previous reports have found that the detection of KRAS mutations with ctDNA in the post-operative setting is a significant prognostic marker for recurrence and survival in patients with pancreatic ductal adenocarcinoma (PDAC) receiving curative surgery [19]. These differences may be due to the extensive neoadjuvant treatment in our study and the relatively small sample size.
The DDR pathway genes such as BRCA1/2, MLH1, and ATM maintain genomic stability and prevent the accumulation of mutations leading to cancer [20]. Platinum-based chemotherapy agents such as oxaliplatin in mFOLFIRINOX act by interfering with DNA replication by forming DNA adducts [21]. Therefore, it has been proposed that tumors with DDR pathway damage may be more sensitive to platinum compounds due to dysfunction in repairing platinum-induced DNA damage. Previous studies of various solid tumors have shown the clinical relevance of this theory [22,23]. As well as other types of tumors, DDR gene mutations may define a subset of patients with pancreatic cancer that may benefit more from platinum-based chemotherapy regimens such as FOLFIRINOX. In a previous study, the presence of germline or somatic DDR gene mutations was associated with better OS in metastatic PDAC patients treated with FOLFIRINOX (hazard ratio, 0.37; 95% CI, 0.15 to 0.94; p=0.04) [24]. In our study, patients with DDR gene mutations identified in plasma ctDNA had better PFS (26.6 months vs. 13.1 months; p=0.009) under neoadjuvant mFOLFIRINOX. Previous studies have investigated DDR gene mutations mainly through tissue samples of resectable or metastatic pancreatic cancer using NGS [25]. Here, we observed similar results to these previous reports using plasma ctDNA, which is obtained less invasively. In addition, this study was conducted on patients with tumors classified as a borderline resectable stage, when the treatment plan is still largely undetermined.
KRAS mutation is one of the most commonly observed oncogene alterations in PDAC. It is critical in initiating and maintaining the disease [26]. Previous studies with ctDNA analysis of patients with PDAC have reported varying KRAS detection rates from ctDNA, ranging from 23% to 49% [27,28]. In our study, we observed the presence of KRAS mutation with plasma ctDNA in about 24% of PDAC patients, similar to previous studies. This is far lower than KRAS detection rates with tissue analysis, where KRAS mutations are expected to be found in more than 80% of patients with pancreatic cancer [26]. Guo et al. [27] have reported KRAS detection rates of 23% from plasma ctDNA, and 74.7% of these patients had KRAS mutations detected in tissue. The detection rate for ctDNA may be lower compared to tissue NGS because pancreatic tumors are generally poorly vascularized [28]. This may also explain the significant association between KRAS alterations detected from plasma at baseline and a poor OS (median 8.5 months vs. not reached, log-rank p=0.003), similar to findings of previous studies [29,30]. Detection of KRAS alterations from plasma at baseline may reflect more shedding of the tumor cells and micrometastases.
This study has several limitations. This was a single-center, retrospective study with a small cohort size and limited validation. Also, the number of patients excluded due to sample contamination was relatively large (n=10 for baseline and n=6 for post-operative), leading to limited results regarding statistical significance. Despite these limitations, our study has strengths in that the patients included in our study were collected from a well-designed and performed phase II clinical trial with homogenous clinical characteristics.
In conclusion, this study investigated the clinical implications of plasma ctDNA analysis of patients with BRPC treated with neoadjuvant mFOLFIRINOX, focusing on the prognostic value of the detection of DDR gene mutations. The detection of DDR gene mutations in plasma ctDNA at baseline was significantly associated with better survival. The detection of KRAS mutations in plasma ctDNA at baseline was significantly associated with worse survival. Additional studies with a larger cohort size are necessary to investigate the precise role of ctDNA and the detection of DDR gene alterations in aiding therapeutic decisions for patients with BRPC in the neoadjuvant setting.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
Notes
Ethical Statement
All procedures in studies involving human participants were performed in accordance with the ethical standards of the Institutional Review Board of Asan Medical Center and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards (IRB approval no. 2016-0010). Patient consent was waived due to the retrospective, anonymized nature of the study.
Author Contributions
Conceived and designed the analysis: Hyung J, Yoo C.
Collected the data: Kim KP, Ryoo BY, Lee SS, Park DH, Song TJ, Hwang DW, Lee JH, Song KB, Kim SC, Hong SM, Yoo C.
Contributed data or analysis tools: Hyung J, Yoo C.
Performed the analysis: Lim DH, Hyung J, Yoo C.
Wrote the paper: Lim DH, Yoon H, Hyung J, Yoo C.
Conflicts of Interest
CY received honoraria from Servier, Bayer, AstraZeneca, Merck Sharp & Dohme, Eisai, Celgene, Bristol Myers Squibb, Debiopharm, Ipsen, Kyowa Kirin, Novartis, Boryung Pharmaceuticals, Merck Serono, Mundipharma, Roche, and Janssen, and received research grants from Servier, Bayer, AstraZeneca, Ono Pharmaceuticals, Celgene, Ipsen, Boryung Pharmaceuticals, Ildong Pharmaceuticals, CKD Pharmaceuticals, and HK inno.N.
Acknowledgments
This study was supported in part by the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea [grant number 2020-IL0018].