Should We Perform Repeated Re-biopsy for the Detection of T790M Mutation?
Article information
Abstract
Purpose
Epidermal growth factor receptor (EGFR) T790M mutations have been detected in the second or third rebiopsy, even if the T790M mutation was not identified in the first rebiopsy. This meta-analysis investigated the EGFR T790M mutation detection rates and its additional advantages with repeated rebiopsies.
Materials and Methods
We searched through the PubMed and EMBASE databases up to June 2022. Studies reporting rebiopsy to identify the EGFR T790M mutation in case of disease progression among patients with advanced non-small cell lung cancer and multiple rebiopsies were included. The quality of the included studies was checked using the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool.
Results
Eight studies meeting the eligibility criteria, reporting 1,031 EGFR mutation–positive patients were selected. The pooled EGFR T790M mutation detection rate of the first and repeated rebiopsies were 0.442 (95% confidence interval [CI], 0.411 to 0.473; I2=84%; p < 0.01) and 0.465 (95% CI, 0.400 to 0.530; I2=69%; p < 0.01), respectively. Overall, the pooled detection rate of EGFR T790M mutation was 0.545 (95% CI, 0.513 to 0.576), which increased by 10.3% with repeated rebiopsies.
Conclusion
This meta-analysis identified that repeated rebiopsy increases the detection rate of EGFR T790M mutation by 10.3%, even if EGFR T790M mutation is not detected in the first rebiopsy. Our results indicate that the spatiotemporal T790M heterogeneity can be overcome with repeated rebiopsy.
Introduction
Lung cancer is the second most diagnosed cancer worldwide and the leading cause of cancer deaths [1]. Patients with advanced non–small-cell lung cancer (NSCLC), harboring the epidermal growth factor receptor (EGFR) mutation, are treated with EGFR–tyrosine kinase inhibitors (TKIs) and develop disease progression on EGFR-TKI therapy after a median of 9–13 months [2]. Several mechanisms of acquired resistance to first- or second-generation EGFR-TKIs have been identified [3], among which the EGFR T790M mutation is the most common and important resistant mutation accounting for 50%–65% [4]. Fortunately, patients with acquired EGFR T790M mutations can be treated with third-generation EGFR-TKIs, such as osimertinib and lazertinib, and have significantly longer progression-free survival (median, 8.1 to 18.5 months) than those on cytotoxic chemotherapy (median, 5.6 months) [2,5]. Therefore, confirming the presence of the EGFR T790M mutation in patients with disease progression to either first- or second-generation EGFR-TKIs is paramount [2].
Tissue rebiopsy for EGFR T790M mutation detection has shown high sensitivity and specificity; however, tissue rebiopsy has limitations, such as invasiveness and difficult tissue acquisition [6]. Therefore, the National Comprehensive Cancer Network (NCCN) recommends initial EGFR testing using plasma samples [7]. Nonetheless, owing to the low sensitivity of liquid biopsy, some patients require additional tissue biopsy [8]. Until now, there are no clear criteria for identifying those who should undergo tissue and liquid rebiopsy. Recently, Kim et al. [9] reported that the accuracy of the plasma test was related to tumor burden. That is, the larger and more extensive the tumor, the more accurate the plasma test.
In addition, previous studies reported that EGFR T790M mutations were detected in the second or third rebiopsy even if they were not identified during the first rebiopsy [8,10–12]. In a recent study by Seto et al. [8], EGFR T790M mutations were confirmed in 5.7% of patients in the second rebiopsy and 12.5% of patients in the third rebiopsy whose EGFR T790M mutation was not confirmed in the first rebiopsy. Repeated rebiopsy appears to be helpful in detecting the EGFR T790M mutation; however, systematic studies of this issue are currently lacking. Therefore, this meta-analysis aimed to investigate the yield of EGFR T790M mutation detection and its additional advantages with repeated rebiopsy.
Materials and Methods
1. Literature search
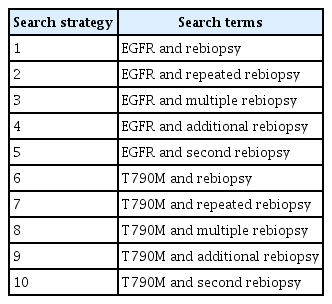
The current meta-analysis was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta Analyses (PRISMA) guidelines [13]. We searched through the PubMed and EMBASE databases for English language articles published until June 2022 using the following search terms: “EGFR” or “T790M” in combination with “rebiopsy,” “repeated rebiopsy,” “multiple rebiopsy,” “additional rebiopsy,” “second rebiopsy” (Table 1).
2. Study selection
Two authors (S.K. and S.H.K.) independently assessed search results using predefined data extraction. Study abstracts were initially reviewed; then, full-text articles were examined for inclusion in this meta-analysis. Discordance was resolved by consensus of three authors (S.K., S.H.K., and J.S.E.). The studies included met all the following criteria: (1) rebiopsy performed to identify EGFR T790M mutation using tissue, cytology, or plasma in the case of disease progression among patients with advanced NSCLC treated with first- or second-generation EGFR-TKI; and (2) rebiopsy performed twice or more. Exclusion criteria were the following: (1) case reports or reviews; (2) non-English language studies; or (3) studies that did not provide sufficient data or were not related to the purpose.
3. Data extraction
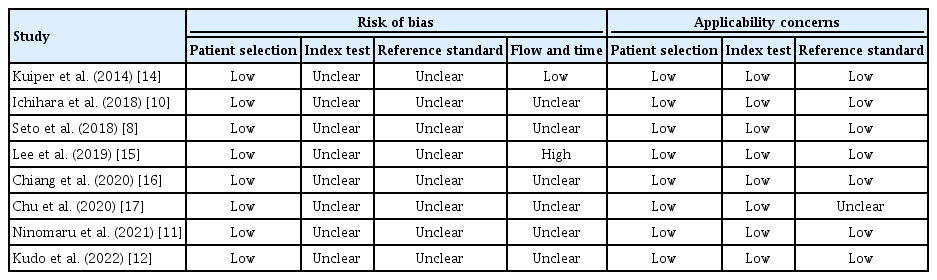
Data were independently extracted by two authors (S.K. and S.H.K.), and discrepancies were resolved by further discussion with the third author (J.S.E.). The following details were extracted from the eligible publications: name of first author, publication year, number of participants, median age, biopsy sites, number of biopsies, biopsy methods, and mutation detection rate. The quality of the included studies was checked using the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool, which assesses the quality of studies by evaluating the four key domains consisting of patient selection, index test, reference standard, and flow of patients [18].
4. Statistical analysis
Statistical analyses were performed using RevMan 5. A fixed-effects model with an inverse variance-weighted method was used to estimate the diagnostic rate of repeated rebiopsy. Study heterogeneity was evaluated using the chi-square statistic test (χ2 test), and quantified by the I2 index [19]. Statistical heterogeneity was considered as significant heterogeneity when the p-value was < 0.01 in the χ2 test, and the I2 index value was > 50%. Publication bias was evaluated by applying a funnel plot together with Egger’s and Begg’s tests [20]. A p-value less than 0.05 was considered statistically significant. Subgroup analysis was performed based on biopsy method (tissue vs. plasma).
Results
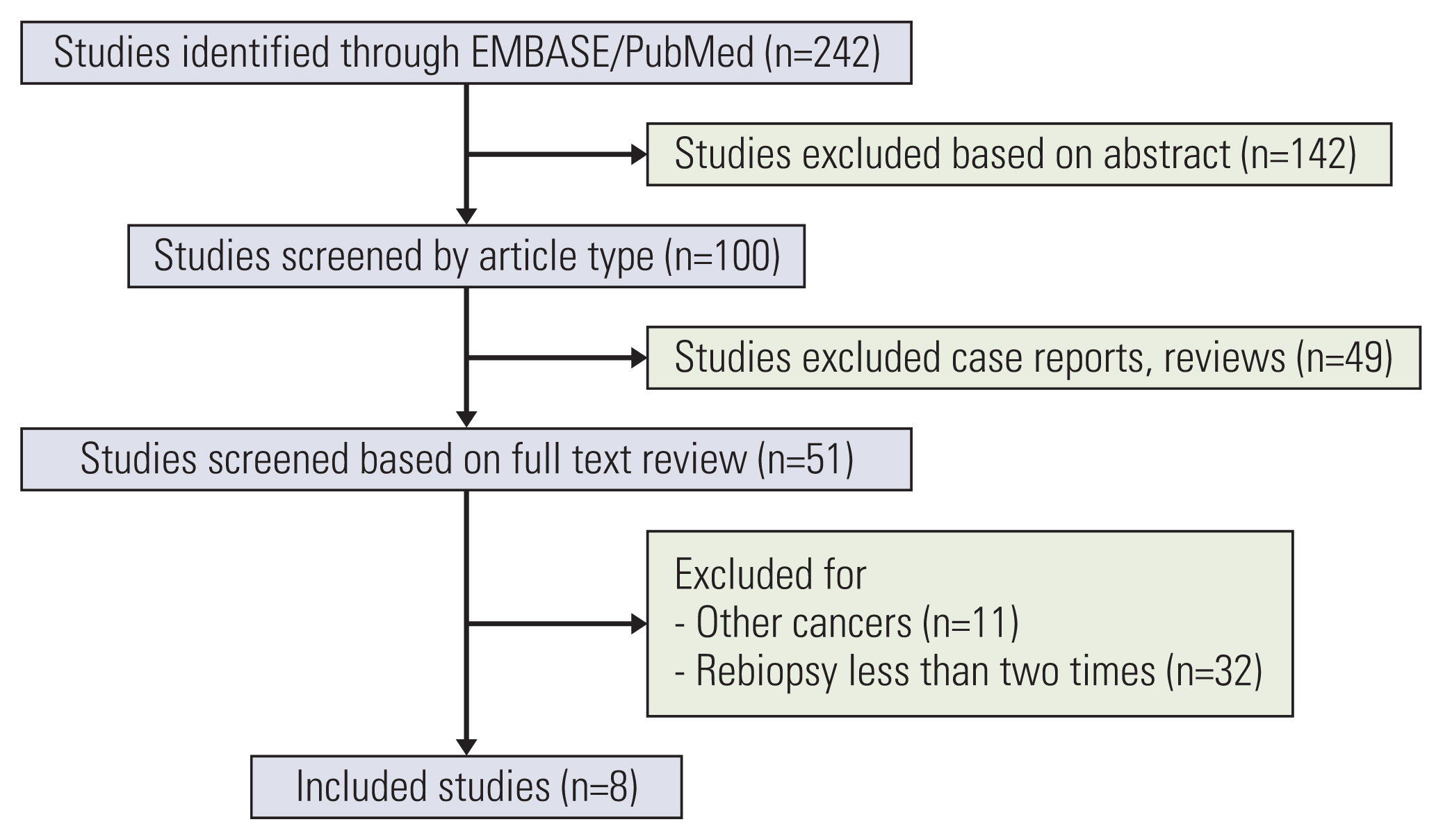
We identified 242 studies from the PubMed and EMBASE databases. After eliminating inapplicable and/or failing to meet the inclusion criteria articles through abstract screening, eight studies were finally selected (Table 2, Fig. 1) [8,10–12,14–17]. As a result, data from 1,031 EGFR mutation–positive patients who reported disease progression after treatment with first- or second-generation EGFR-TKI were analyzed. The first biopsy performed after disease progression was referred to as the first rebiopsy, and the additional biopsy, when EGFR T790M mutation was not detected in the first rebiopsy, was referred to as the repeated rebiopsy.
Table 3 provides a quality assessment of all included studies based on QUADAS-2. The overall analysis showed good performance in the patient selection and index test criteria. However, it showed poor performance in the flow and time criteria. The funnel plot was symmetric (Fig. 2), with both Egger’s (p=0.573) and Begg’s tests (p=0.823) showing insignificant p-values, indicating an absence of publication bias.
1. EGFR T790M detection rates
In the first rebiopsy, EGFR T790M mutation was found in 448 of 1,031 patients, and the pooled EGFR T790M mutation detection rate was 0.442 (95% confidence interval [CI], 0.411 to 0.473; I2=84%; p < 0.01). Of the 583 patients without EGFR T790M mutation in the first rebiopsy, 248 (42.5%) underwent repeated rebiopsies, and the pooled detection rate of EGFR T790M mutation of repeated rebiopsy was 0.465 (95% CI, 0.400 to 0.530; I2=69%; p < 0.01). Overall, the pooled detection rate of EGFR T790M mutation was 0.545 (95% CI, 0.513 to 0.576; I2=91%; p < 0.01), which increased by 10.3% with repeated rebiopsies (Fig. 3).
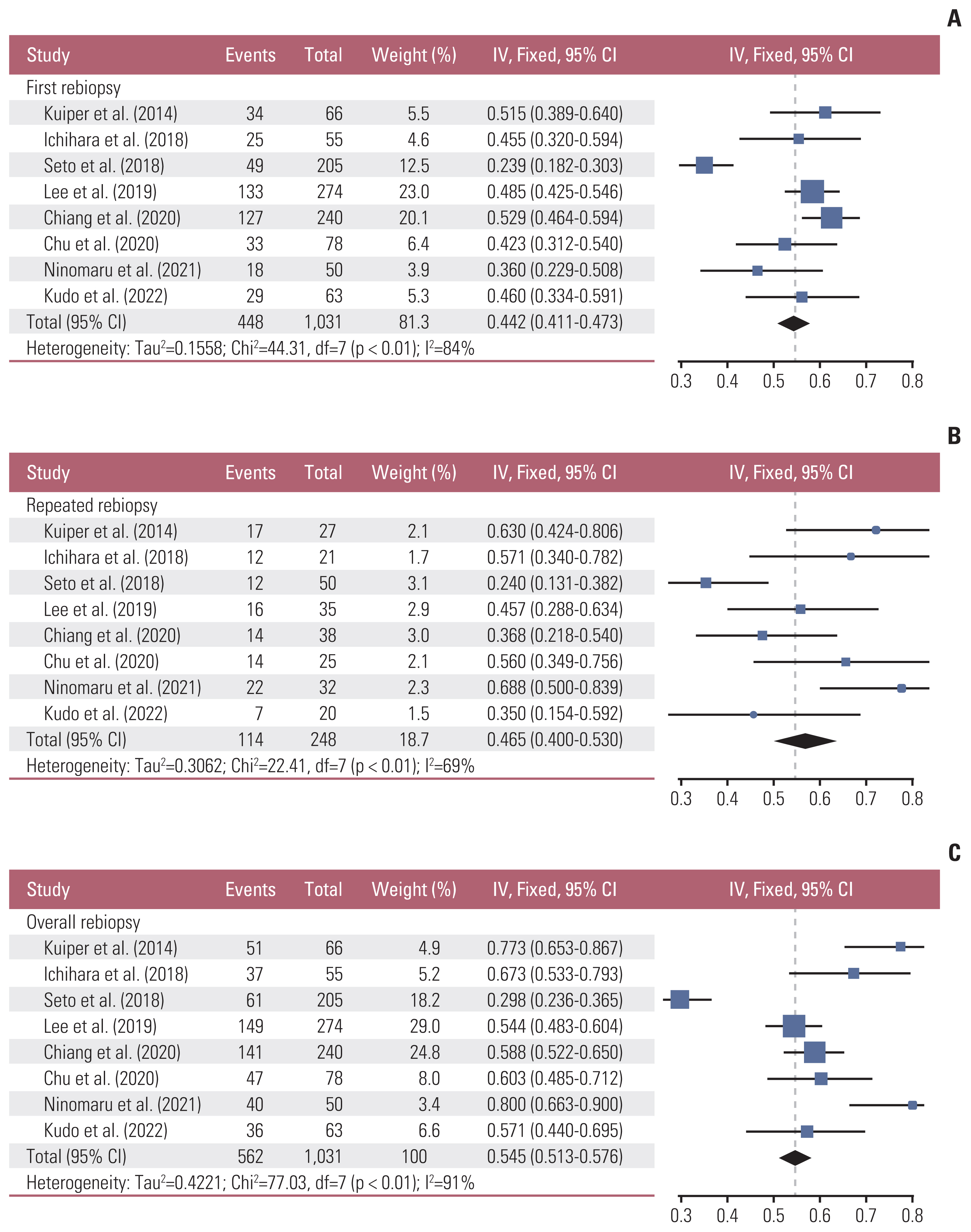
Forest plot of epidermal growth factor receptor (EGFR) T790M mutation detection rate [8,10–12,14–17]. (A) EGFR T790M mutation detection rate in the first rebiopsy. (B) EGFR T790M mutation detection rate in the repeated rebiopsy. (C) Pooled EGFR T790M mutation detection rate of the first and repeated rebiopies. CI, confidence interval.
In some cases, the EGFR T790M mutation was negative in liquid biopsy but turned positive when repeated rebiopsy was performed using tissue samples, and vice versa. Seto et al. [8] reported that among 46 patients with negative EGFR T790M mutation based on tissue or cytology samples, two (4.3%) were positive for EGFR T790M mutation when repeated rebiopsy was performed using plasma samples [8]. The opposite was observed in five out of 110 patients (4.5%).
2. Subgroup analyses
In the first rebiopsy using tissue or cytology, the pooled EGFR T790M mutation detection rate was 0.380 (95% CI, 0.297 to 0.471; I2=50%; p=0.157) [8,12]. However, in the repeated rebiopsy using tissue or cytology, the pooled EGFR T790M mutation detection rate was 0.260 (95% CI, 0.180 to 0.358; I2=0%; p=0.746) [8,12,16]. Moreover, the pooled EGFR T790M mutation detection rate was 0.226 (95% CI, 0.164 to 0.302; I2=81%; p=0.023) in the first liquid rebiopsy [8,12] and was 0.310 (95% CI, 0.150 to 0.534; I2=71%; p=0.030) in the repeated liquid rebiopsy [8,12,16].
Discussion
This is the first meta-analysis to evaluate the efficacy of repeated rebiopsy for detecting EGFR T790M mutation. In this report, the overall EGFR T790M mutation detection rate was 54.5% and the repeated rebiopsies increased the pooled detection rate by 10.3%. Our analyses suggest that even though the EGFR T790M mutation was not detected in the first rebiopsy, repeated rebiopsy could help detect the EGFR T790M mutation.
Individual tumors have variations in genetic diversity over time, and there is an unequal distribution of genetic diversity in different sites or within a single tumor site, called spatiotemporal heterogeneity [21]. Tumor heterogeneity plays an important role in acquired resistance to targeted therapy in patients with advanced NSCLC [22]. Consequently, evaluating the state of evolutional changes in the somatic mutations through iterative analyses is important, which can lead to appropriate treatment agent selection. Furthermore, the EGFR T790M–positive strain may not be detected at the first time due to the spatiotemporal heterogeneity of tumors, and several studies have consistently reported the importance of iterative evaluation through repeated rebiopsy [11,21]. In the present study, repeated rebiopsy increased the overall EGFR T790M mutation detection rate by 10.3%, which suggests that the spatiotemporal heterogeneity of EGFR T790M mutation can be overcome with repeated rebiopsy. In particular, repeated liquid rebiopsy should be preferentially performed because it is simple, convenient, non-invasive, and can overcome spatial heterogeneity.
Kim et al. [23] reported that the smaller the lung lesion, the higher the detection rate of EGFR T790M mutation. Recently, Hong et al. [24] found that lymph node sampling using endobronchial ultrasound-guided transbronchial needle aspiration was more appropriate for EGFR T790M mutation detection than lung biopsy using radial probe endobronchial ultrasound. In addition, Oxnard et al. [25] demonstrated that the EGFR T790M mutation detection rate was higher in the lung, pleura, and lymph nodes than in distant sites. Although there has been controversy regarding the favorable anatomical site of EGFR T790M mutation development and detection, the data suggest that spatial heterogeneity in the EGFR T790M mutation development is evident. Moreover, Lin et al. [26] recently reported that EGFR T790M mutation was found in 17% of advanced NSCLC patients who underwent salvage surgery after EGFR-TKI treatment (mean duration of EGFR-TKI treatment before salvage surgery=134 days), indicating that EGFR T790M mutation develops gradually at the beginning of EGFR-TKI treatment, so-called temporal heterogeneity. Although there are no gold standards for selecting the biopsy method, site, and timing, active rebiopsy is required whenever disease progression is confirmed to overcome spatial and/or temporal heterogeneity.
In the subgroup analysis, the detection rate of the first tissue rebiopsy for the EGFR T790M mutation was 38%, whereas the yield of repeated tissue rebiopsy decreased to 26%. On the contrary, the EGFR T790M mutation detection rate of repeated liquid rebiopsy was higher than that of the first liquid rebiopsy (22.6% vs. 31%). Notably, the detection rate of EGFR T790M mutation of repeated liquid rebiopsy was numerically higher than that of repeated tissue rebiopsy (31% vs. 26%). According to previous data, the sensitivity of EGFR T790M mutation detection in tissue samples is higher than in plasma samples [6]. Generally, tissue acquisition is not always easy in advanced lung cancer patients, especially those already heavily treated [8]. Moreover, a previous study demonstrated that tissue sampling is possible in only 55% of patients [27]. Most patients with relapse or progression have a poor general condition, making it difficult to acquire tissue or cytology samples. Therefore, there may be a possible tendency to prefer liquid biopsy in repeated rebiopsy and repeated tissue rebiopsy may be determined in a limited number of study subjects.
There are advantages and disadvantages associated with repeated rebiopsy. First, there are concerns regarding the procedure-related complications developed after repeated rebiopsy. However, until now, life-threatening complications of repeated rebiopsy have not been reported. In addition, there is no difference in the complication rate between the first tissue rebiopsy and the repeated tissue rebiopsy. Chu et al. [17] reported that 11% of patients experienced complications in the first rebiopsy and 9% in repeated rebiopsy. On the contrary, the benefits of repeated rebiopsy are also evident. For example, clinical outcomes, such as the objective response rate, disease control rate, and progression-free survival, with osimertinib were not significantly different between patients with EGFR T790M mutation at the first rebiopsy and patients with EGFR T790M mutation at the repeated rebiopsy (Table 4) [10–12,16].
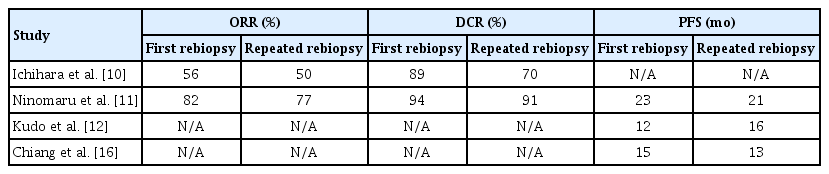
Comparisons of clinical outcomes after osimertinib treatment between patients with T790M at the first rebiopsy and patients with T790M at the repeated rebiopsy
This study has several limitations. First, the number of studies and patients was relatively small. Second, only 42.5% of EGFR T790M mutation-negative patients in the first rebiopsy underwent repeated rebiopsy. A more aggressive repeated rebiopsy may increase the overall EGFR T790M mutation detection rate. Third, the possible differences in the priorities of tissue or plasma samples by each research institution were not reflected. Fourth, this meta-analysis did not reflect that each study had different expertized doctors and medical resources, which may also affect the yield of tissue rebiopsy. Fifth, there are several PCR-based diagnostic methods for detecting the EGFR T790M mutation, and each test has different sensitivity and specificity for EGFR T790M detection [28,29]. However, in the literature reviewed in this meta-analysis, the data were insufficient to compare the EGFR T790M mutation detection rate among different methods and kits. Sixth, most repeated rebiopsies in this study were performed at the time of disease progression. Until now, there are no established definitive data on the optimal timing for repeated rebiopsy; hence, further research is needed in this regard. Finally, owing to the heterogeneity of the included studies, the detection rates of EGFR T790M mutations according to rebiopsy sites (primary vs. intrathoracic or extrathoracic metastatic lesions) could not be compared, which might limit the interpretation of our study results.
In conclusion, this study identified that repeated rebiopsy increases the detection rate of EGFR T790M mutation in patients with advanced NSCLC, even if EGFR T790M mutation was not detected in the first rebiopsy. Our results indicate that the spatiotemporal heterogeneity of EGFR T790M mutation in individual patients with EGFR-mutant NSCLC after acquired resistance to EGFR-TKI can be overcome with repeated rebiopsy.
Notes
Author Contributions
Conceived and designed the analysis: Kim S, Kim SH, Kim MH, Lee MK, Eom JS.
Collected the data: Kim S, Kim SH, Kim MH, Lee MK, Eom JS.
Contributed data or analysis tools: Kim S, Kim SH, Kim MH, Lee MK, Eom JS.
Performed the analysis: Kim J, Eom JS.
Wrote the paper: Kim S, Kim SH, Eom JS.
Conflicts of Interest
Eom JS received speaker fees from Yuhan, Boehringer Ingelheim, Amgen, AstraZeneca, Olympus, and Erbe Corporations as an invited speaker at academic medical meetings. Eom JS reports grants from Boehringer Ingelheim. Kim SH received speaker fees from AstraZeneca. Kim S has no conflict of interest directly relevant to the content of this article.
Acknowledgments
We thank the Department of Biostatistics, Biomedical Research Institute, and Pusan National University Hospital. This work was supported by a clinical research grant from the Pusan National University Hospital in 2023. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (grant No. NRF-2021R1F1A1047622).