Use of a High-Throughput Genotyping Platform (OncoMap) for RAS Mutational Analysis to Predict Cetuximab Efficacy in Patients with Metastatic Colorectal Cancer
Article information
Abstract
Purpose
Cetuximab demonstrates improved efficacy outcomes in patients with metastatic colorectal cancer (mCRC) harboring wild-type KRAS exon 2. Resistance to cetuximab is mediated by activating less frequent mutations in the RAS genes beyond KRAS exon 2. We performed extended RAS Mutational analysis using a high-throughput genotyping platform (OncoMap) and evaluated extended RAS analysis for predicting cetuximab efficacy in patients harboring wild-type KRAS exon 2 tumors following Sanger sequencing.
Materials and Methods
Extended RAS analysis was performed on 227 wild-type KRAS exon 2 mCRC patients who received cetuximab as salvage treatment using OncoMap ver. 4.0. Targeted genes included exon 2, exon 3, and exon 4, both in KRAS and NRAS, and included BRAF exon 15. We assessed efficacy by the new RAS mutation status.
Results
The OncoMap detected 57 additional mutations (25.1%): 25 (11%) in KRAS exon 2 and 32 (14.1%) beyond KRAS exon 2. Survival differences were observed after dividing patients into the wild-type RAS group (n=170) and mutant RAS group (n=57) using OncoMap. Progression-free survival was 4.8 months versus 1.8 months (hazard ratio [HR], 0.44; 95% confidence interval [CI], 0.32 to 0.61), and overall survival was 11.9 months versus 8.4 months (HR, 0.65; 95% CI, 0.47 to 0.88).
Conclusion
Sanger sequencing is not sufficient for selecting candidates for cetuximab treatment. High-throughput extended RAS genotyping is a feasible approach for this purpose and identifies patients who might benefit from cetuximab treatment.
Introduction
Colorectal cancer is the third most common cancer and the fourth leading cause of cancer-related death in Korea [1]. As the development of chemotherapeutic agents improved treatment outcomes, the median overall survival (OS) of metastatic colorectal cancer (mCRC) patients treated with chemotherapy higher to over 20 months. The efficacy of anti–epidermal growth factor receptor (EGFR) monoclonal antibodies (mAbs) is one important development. Cetuximab— a chimeric monoclonal antibody that targets EGFR—demonstrated improved efficacy outcomes in mCRC patients harboring wild-type KRAS exon 2 through all treatment continuums, from the first-line to salvage-line treatments [2,3].
RAS genes are common oncogenes in human cancer and present in 30%-40% of colorectal cancers [4]. Of the three major isoforms of RAS—including KRAS, NRAS, and HRAS—mutant KRAS exon 2 is the most prevalent RAS mutation in colorectal cancer. Additional RAS mutations beyond the KRAS exon 2 can be found in 15%-27% of tumors harboring wild-type KRAS exon 2, and they reduce the efficacy of anti-EGFR mAbs–based treatment [5-8]. In the FIRE-3 trial (folinic acid, 5-fluorouracil and irinotecan [FOLFIRI] plus cetuximab vs. FOLFIRI plus bevacizumab as first-line treatment for patients with mCRC), OS in RAS wild-type subgroup after extended RAS testing was higher compared with patients with mCRC with the wild-type KRAS exon 2 [5]. The PRIME (The Panitumumab Randomized Trial in Combination with Chemotherapy for Metastatic Colorectal Cancer to Determine Efficacy) and PEAK (panitumumab plus 5-fluorouracil, folinic acid and oxaliplatin [FOLFOX] or bevacizumab plus FOLFOX in patients with mCRC) trials added weight to this argument [6,7]. Extended RAS testing beyond KRAS exon 2 is accepted and recommended in various countries, including the United Kingdom, France, Japan, and United States, before treatment with anti-EGFR mAbs. The current recommendations are derived from retrospective subgroup analyses of first-line trials [6,9,10]; however, there are few trials that report the impact of extended RAS testing under salvage-line settings in Asian patients.
There are several methods for RAS testing, and their sensitivities vary. Sanger sequencing and real-time polymerase chain reaction (PCR) are approved for analyzing KRAS exon 2 mutations in Korea. Sanger sequencing for RAS mutations beyond KRAS exon 2 was also recently approved but was not used before 2015. Although Sanger sequencing is widely used in clinical practice, this test has a low sensitivity with about 20% of detection limit [11]. In addition, the test is laborious and time-consuming. Real-time PCR is more sensitive than Sanger sequencing; however, it is not approved for testing extended RAS beyond KRAS exon 2 in Korea. OncoMap is a technology that can detect mutations in cancer-related genes with speed, accuracy, and a sensitivity with about 5% of detection limit using the mass spectrometer [12]. OncoMap can detect specific mutations with the following process. First, the target DNA is amplified. Second, single base extension is performed. After the single base extension reaction, small DNA products that have unique mass value according to mutation are generated. These differences are measured by mass spectrometer.
The first aim of our study was to evaluate if the high-throughput genotyping platform—OncoMap—can detect RAS mutations accurately in KRAS exon 2 wild-type patients determined by Sanger sequencing. The second aim was to evaluate the impact of extended RAS in chemotherapyrefractory mCRC patients treated with cetuximab.
Materials and Methods
1. Study population
To evaluate the impact of extended RAS testing, we identified wild-type KRAS exon 2 patients who were diagnosed with histologically confirmed mCRC. Three hundred and sixteen consecutive patients were treated at our hospital between December 2003 and June 2013 with cetuximab as third-line or later treatments after oxaliplatin, irinotecan, and fluoropyrimidines failed. Patients were excluded if they had tumor tissue inappropriate for further genetic analyses (n=89). Two hundred and twenty-seven patients were finally included in our study population. An objective response analysis was available for 202 patients with measurable disease. The Institutional Review Board of Asan Medical Center approved this study.
2. Tumor tissue sampling and mutational analysis
Formalin-fixed paraffin embedded (FFPE) tissues of primary or metastatic lesions were used for the genetic analyses. The FFPE tissue blocks were retrieved from archives, reviewed by a pathologist, and the tumor portion was marked and cut for genetic analyses. Genomic DNA was extracted from FFPE tissue using the QIAamp DNA Tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The extracted genomic DNA was then analyzed using the currently available OncoMap ver. 4.0 [13]; 471 mutations in 41 cancer-related oncogenes can be detected using the OM_V4_Core format. For this study, the OncoMap-Colon Panel included 11 hot spots: codons 12 and 13 (exon 2), codon 61 (exon 3), and codons 117 and 146 (exon 4), both in the KRAS and NRAS oncogenes, and exon 15 in the BRAF oncogenes.
3. Statistical analysis
The primary study objective was to determine the frequency of additional RAS mutations that were detected by the high-throughput genotyping platform (OncoMap) in mCRC patients with wild-type KRAS exon 2 documented by Sanger sequencing. The secondary objective was to evaluate whether more sensitive RAS testing would predict the efficacy of cetuximab treatment. Objective response rate (ORR) was determined using Response Evaluation Criteria in Solid Tumors ver. 1.1. Progression-free survival (PFS) was defined as the time from the administration of cetuximab to disease progression or death-related to disease, and OS as the time to death from any cause.
All statistical analyses were explorative. Fisher exact test was used to compare the ORR between the RAS groups (wild-type vs. mutant). In addition the odds-ratio and 95% confidence intervals were calculated for ORR by RAS status. The Kaplan-Meier method was used to estimate PFS and OS, and log-rank test was applied to compare both endpoints by RAS status. The hazard ratio (HR) and corresponding 95% confidences intervals for PFS and OS were calculated using univariate Cox proportional hazards methods. Two-tailed null hypotheses of no difference were rejected if p-values were less than 0.05, or, equivalently, if the 95% confidence intervals (CIs) of risk point estimates excluded 1. No alpha adjustment was applied for multiple significance testing. All data were analyzed using the SPSS ver. 21.0 (IBM Corp., Armonk, NY).
Results
1. Baseline characteristics of the study subjects
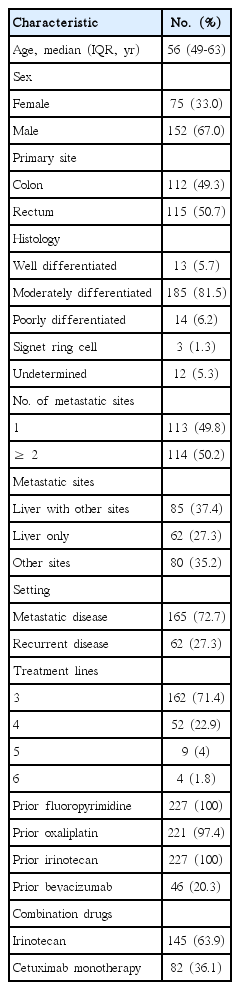
The clinical characteristics of the 227 study patients are presented in Table 1. The median age was 56 years, and 152 patients (67%) were male. Of these 227 patients, 165 patients (72.7%) had metastatic disease at diagnosis, whereas 62 patients (27.3%) had recurrent disease. The most common site of metastasis was the liver (64.8%). All patients had been treated with fluorouracil and irinotecan. There were no significant differences in any baseline characteristics by RAS mutation status. The prior treatment period from the date of first-line chemotherapy to the date of cetuximab administration, was 17.8 months in the wild-type RAS group and 17.4 months in the mutant RAS group. The median duration of cetuximab treatment was 13 weeks (interquartile range, 6 to 24.9 weeks).
2. Additional RAS mutations
All 227 study patients were documented with tumors harboring wild-type KRAS exon 2 by Sanger sequencing. After reanalysis with OncoMap, 25.1% of these patients (57 of 227) were identified as harboring additional RAS mutations (i.e., any mutant RAS group) (Table 2). Of note, 11% of the patients (25 of 227) had a mutated KRAS exon 2, which was not detected by Sanger sequencing. Beyond KRAS exon 2, 14.1% of patients (32 of 227) had mutations in KRAS exons 3 or 4, or NRAS exons 2, 3, or 4 (Table 2). Of the 202 patients with confirmed wild-type KRAS exon 2, 15.8% (32 of 202) had RAS mutations beyond KRAS exon 2. The detailed frequencies of the RAS mutations are listed in Table 2.
3. Clinical outcomes according to RAS mutations
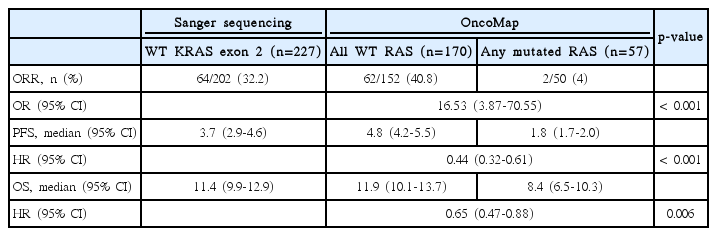
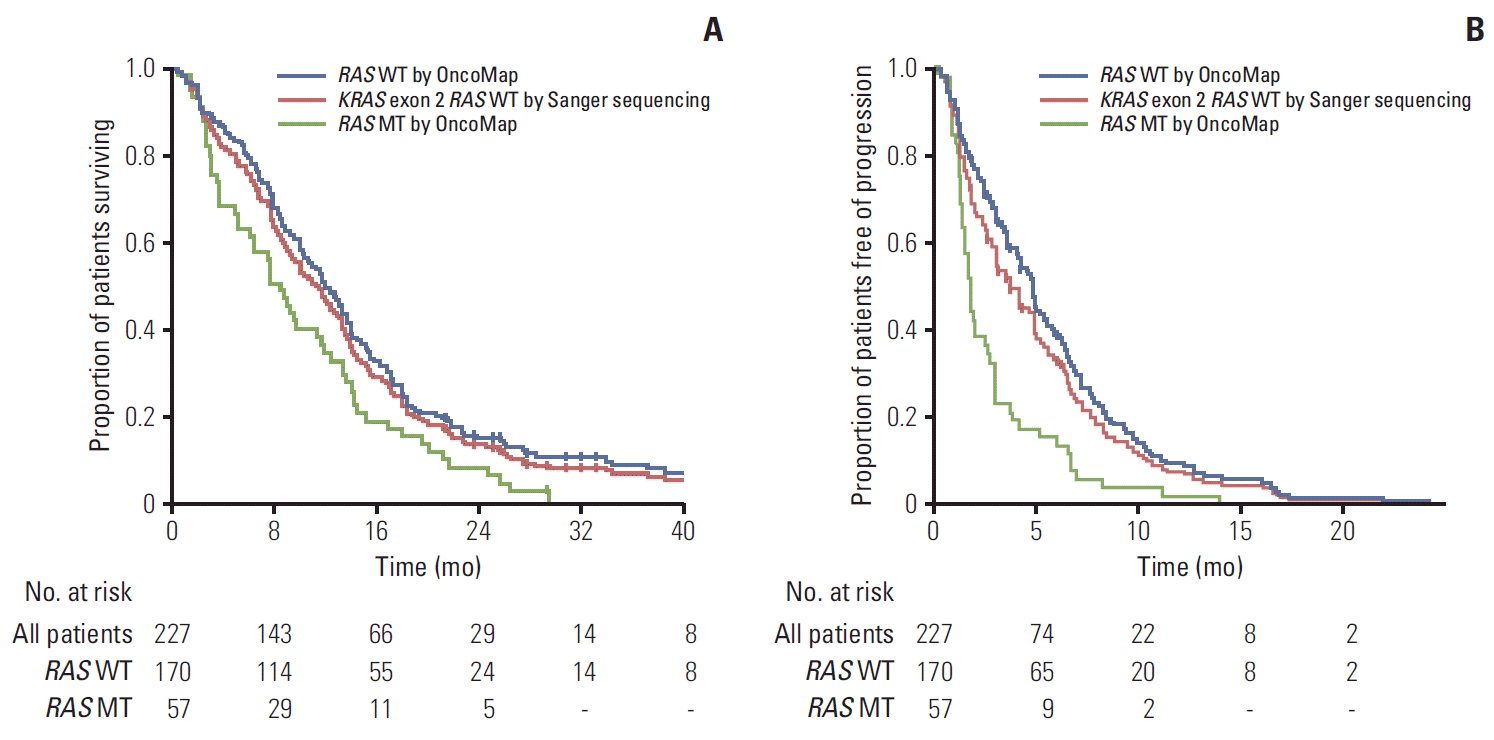
The treatment outcomes are presented in Table 3. Among the 227 patients known to harbor wild-type KRAS exon 2, as determined by Sanger sequencing, the PFS was 3.7 months(95% CI, 2.9 to 4.6) and the median OS was 11.4 months (95% CI, 9.9 to 12.9). Objective responses were observed in 64 of the 202 patients evaluated for a response (31.7%). The odds ratio was 16.53 (95% CI, 3.87 to 70.55; p < 0.001). Separation of the Kaplan-Meier curve was observed between groups (Fig. 1) by the new RAS Mutational status. The median OS was 11.9 months (95% CI, 10.1 to 13.7) in the wild-type RAS group according to the OncoMap and 8.4 months (95% CI, 6.5 to 10.3) in the mutant RAS group (p=0.006). The PFS differed significantly between groups (4.8 months [95% CI, 4.2 to 5.5] vs. 1.8 months [95% CI, 1.7 to 2.0]; p < 0.001). Of the 57 patients with RAS mutations detected by OncoMap, 55 patients’ disease progressed following cetuximab therapy.
4. BRAF mutation
The BRAF V600E mutation was detected in 6.2% of the study patients (14 of 227). BRAF mutations were found only in individuals harboring wild-type RAS tumors. Among the 170 patients with wild-type RAS tumors according to the OncoMap, the OS was 12.8 months (95% CI, 11.4 to 14.2) in the wild-type BRAF group and 2.3 months (95% CI, 0 to 4.9) in the mutant BRAF group. The PFS in the non-mutated BRAF patients was significantly longer than in the mutant BRAF patients (5.0 months [95% CI, 4.3 to 5.6] vs. 1.1 months [95% CI, 0.4 to 1.7]; HR, 0.10; 95% CI, 0.05 to 0.18). None of the 12 evaluable patients in the mutant BRAF group showed a response to cetuximab.
5. Survival in patients with wild-type RAS and wild-type BRAF mCRC
In patients with RAS WT and BRAF wild-type mCRC (all wild-type group, n=156), PFS was 5.0 months (95% CI, 4.3 to 5.6) compared with 1.6 months (95% CI, 1.3 to 2.0) in mutant RAS or mutant BRAF patients (any mutant group, n=71). OS was 12.8 months (95% CI, 11.4 to 14.2) and 7.6 months (95% CI, 5.2 to 10.0) in the all wild-type group and any mutant group, respectively.
Discussion
We found that 14.1% of additional RAS mutations in Korean mCRC populations exist beyond KRAS exon 2. To the best of our knowledge, this is the first study to report a need for extended RAS testing in Asian mCRC patients treated with cetuximab. This is consistent with reports in Western populations in a variety of settings, from first-line to salvage therapies [5,6,8,9,14]. We earlier found frequency of KRAS exon 2 mutations in a Korean case series to be similar to the Western studies [15]. The frequency of extended RAS mutations in our current case series likewise does not differ from Western populations.
Another interesting finding of our current study was that we detected additional KRAS exon 2 mutations (11% of the total) that could not be found using Sanger sequencing, a technique that is widely used in general clinical practice. Furthermore, the patients with mutated RAS according to OncoMap showed significantly worse outcome compared to wild-type RAS patients under cetuximab treatment. Atreya et al. [14] suggest that additional KRAS mutations could be detected in 20%-30% of wild-type KRAS patients using routine testing if the tumors with rare mutant KRAS clones are examined using a higher sensitivity assay. Trials such as the PRIME, FIRE-3, and the CRYSTAL trial did not report the additional detection of the KRAS exon 2 mutations because they used a sensitive method for detecting KRAS exon 2 mutations (real-time PCR and pyrosequencing).
Among the patients in the FIRE-3 trial treated with first-line cetuximab-FOLFIRI chemotherapy, re-analysis of survival after extended RAS testing showed that median OS in wild-type RAS patients is longer than wild-type KRAS patients (33.1 months vs. 28.7 months, respectively) [5]. Among the patients who received first-line panitumumab-FOLFOX in the PRIME trial, PFS (9.6 to 10.1 months) and OS (23.9 to 26 months) improved according to the results of extended RAS testing [6]. However, there have been few studies that investigated the role of extend RAS testing under salvage settings in mCRC patients. Our present data suggest that the benefit of extended RAS testing could be applied to chemotherapy-refractory mCRC patients treated with cetuximab.
OncoMap may be able to overcome the weaknesses of Sanger sequencing. First, OncoMap is a more sensitive method. Furthermore, there are many advantages of OncoMap, such lower tissue requirement, shorter turnaround time, and automated methodology. In addition, it can test other mutations of interest at the same time. There is always debate however, as to whether more accurate tests are always better. The different analytical techniques used to evaluate RAS Mutational status have not influenced the predictive value of RAS mutations [16]. Moreover, some researchers report that patients with tumors containing rare mutant RAS cells might clinically benefit from anti-EGFR mAbs [9,14]. Further research is needed to define the cutoff values for the detection limit of RAS mutations.
In our study, two patients responded to treatment despite having mutant KRAS tumors, detected by OncoMap. They had KRAS mutations (Q61H in exon 3 and G12D in exon 2) and were treated with cetuximab in combination with irinotecan. They lived for 6.9 months and 11.1 months with out cancer progression. In the case with G12D in exon 2, the tumor with small RAS clones might be responding to cetuximab/irinotecan chemotherapy, because OncoMap is more sensitive than Sanger sequencing.
The clinical usefulness of identifying the BRAF mutation for anti-EGFR mAbs therapy is unclear. In the pooled analysis of CRYSTAL (FOLFIRI plus cetuximab in mCRC) and OPUS (cetuximab plus FOLFOX as first-line treatment for mCRC), the BRAF mutation was a poor prognostic marker in patients with wild-type KRAS mCRC who were treated with cetuximab in combination with chemotherapy as the first-line treatment [17]. In the retrospective analysis of the earlier PRIME trial, the survival outcomes in mutant BRAF patients (n=24) without RAS mutations were inferior to those of wild-type BRAF patients (PFS, 6.1 months [95% CI, 3.7 to 10.7] vs. 10.8 months [95% CI, 9.4 to 12.4]; OS, 10.5 months [95% CI, 6.4 to 18.9] vs. 28.3 months [95% CI, 23.7 to not evaluated]) [6]. In chemotherapy refractory settings, the role of the BRAF mutation in anti-EGFR mAbs therapy is not wellestablished due to the small number of mutant BRAF patients [18,19]. According to our current data, although our sample size was small, the OS and PFS in mutant BRAF patients (n=14) were significantly less than the wild-type BRAF patients.
This study had some notable limitations. First, this was a retrospective analysis from a single center. Second, there was no control group. However, this represents the first study to investigate the frequency and impact of RAS mutations beyond KRAS exon 2 in an Asian population. Our data suggest that false-negative results could occur in routine clinical practice using Sanger sequencing, which has been an approved methodology that is required for extended RAS testing.
Conclusion
In conclusion, Sanger sequencing is not sufficient for selecting candidates for cetuximab treatment. High throughput extended RAS genotyping is feasible and identifies patients who might benefit from cetuximab treatment.
Notes
This study was supported in part by Merck Ltd.
Acknowledgements
The Republic of Korea, with additional support from the Korea Health 21 R&D project, Ministry of Health and Welfare and Family Affairs, Republic Korea (HI06C0868), the National R&D Program for Cancer Control, Ministry of Health and Welfare, Republic of Korea (1420030), and the Asan Institute for Life Sciences, Seoul, Republic of Korea (2015-0753).