An Association Study of Polymorphisms in JAK3 Gene with Lung Cancer in the Korean Population
Article information
Abstract
Purpose
The genetic alteration of the janus kinases (JAKs), non-receptor tyrosine kinase, is related to the development of human cancers. However, little is known about how the sequence variation of JAK3 contributes to the development of lung cancer. This study investigated whether polymorphisms at the promoter region of the JAK3 gene are associated with the risk of lung cancer in the Korean population.
Materials and Methods
A total of 819 subjects, including 409 lung cancer patients and 410 healthy controls were recruited. The SNaPshot assay and polymerase chain reaction-restriction fragment length polymorphism analysis were used, and logistic regression analyses were performed to characterize the association between polymorphisms of JAK3 and lung cancer risk.
Results
Three polymorphisms (-672 G>A, +64 A>G and +227 G>A) of JAK3 were analyzed for large-scale genotyping (n=819). Statistical analyses revealed that polymorphisms and haplotypes in the JAK3 gene were not significantly associated with lung cancer.
Conclusion
JAK3 gene was not significantly associated with the risk of lung cancer in the Korean population.
Introduction
Lung cancer is the leading cause of cancer-related death, accounting for one third of all deaths from cancer worldwide [1,2] and ranks the second highest in incidence in Korea [3]. Lung cancer therapies rarely cure, and the overall 5-year survival rate is still only 15% [2]. Moreover, lung cancer is often diagnosed after the appearance of clinical symptoms, which may be due to primary disease, metastasis or formation of neoplasm [4]. Even though the necessity to reduce the incidence of lung cancer and improve the poor current outcome is very important, the molecular mechanisms for lung cancer development have not been well characterized.
Tyrosine kinase (TK) growth factor receptors on the cell membrane are good targets in cancer therapy because signals from these receptors promote cell growth and survival [5]. They play a critical role in a wide range of biological processes, including embryonic development, organism growth, angiogenesis, synaptic plasticity and oncogenesis [6]. Janus tyrosine kinases (JAKs) are one of eleven mammalian non-receptor TK families that are essential mediators of cellular signaling through cytokine receptors. JAK activates signal transducers and activators of transcription (STAT) factors to dimerize and translocate into the nucleus, which will initiate the transactivation of target genes. This pathway is crucial to hematopoiesis, immune response, oncogenesis, and proliferation [7,8]. The association of JAK with cell growth and proliferation, and its critical role in tumorigenesis of cancer has been investigated previously [9,10]. Furthermore, JAK genes are often mutated in human cancers, suggesting that JAK3 mutations may to be functional and contribute to cancer development including leukemia, breast, and lung cancer [11,12]. Also, in human colorectal cancer, JAK3 expression had a significant association with tumor differentiation, pathological T-stage, and tumor-node-metastasis (TNM) stage [13]. It was reported that the JAK3/STAT3 signaling pathway plays an important role in the pathogenesis of human colon cancer by promoting cell survival and counteracting apoptotic cell death [13,14].
Despite its potential roles in cancer development, there are no comprehensive genetic studies to date on the correlation between JAK3 and lung cancer incidence. In this study, we evaluated the relationship between JAK3 polymorphisms and clinic pathological parameters in Korean lung cancer patients by analyzing genotypes and haplotypes. Our data showed that JAK3 polymorphisms and haplotypes are not significantly associated with lung cancer risk in the Korean population.
Materials and Methods
1. Subjects
Blood samples were collected between August 2001 and June 2006 from 819 subjects, including 409 lung cancer patients and 410 healthy controls. Lung cancer patients were recruited from the patient pool at the Genomic Research Center for Lung and Breast/Ovarian Cancer and Inha University Medical Center. On the other hand, control subjects were randomly selected from the Cardiovascular Genome Center, Genomic Research Center for Allergy and Respiratory disease and Keimyung University Dongsan Medical Center. The histological classification and staging of all patients was performed by pathological evaluation and the clinical or pathological stages of lung cancer at the time of diagnosis were determined by reviewing the medical records based on the TNM system.
Each patient and control subject completed detailed questionnaires covering diet, smoking status, drinking status, lifestyle and medical history, with assistance from a trained interviewer. For the smoking status of the subjects, any subject who reported smoking at least once on a daily basis was considered a smoker for the purpose of this study. All study subjects provided written consent, and were approved for the study protocol by the Institutional Review Board.
2. DNA isolation and genotyping
Total genomic DNA was extracted using the PUREGENE blood DNA purification system (Gentra, Minneapolis, MN), according to the manufacturer s instructions. Purified genomic DNA was eluted in 50 µL elution buffer and 50 ng of DNA was used for polymerase chain reaction (PCR). After initial genotyping among 24 randomly selected samples from lung cancer patients, we focused on polymorphisms in the JAK3 promoter region. We analyzed the region spanning 2 Kb upstream from the translation initiation site (primers shown in Appendix 1). The positioning of polymorphisms, primer and probe designs relative to the transcriptional start site of JAK3 were based on the GenBank sequence (accession no. NT_011295). Single base extension was performed using gene-specific primers according to the manufacturer s protocol (ABI Prism SNaPshot multiplex system, PE Applied Biosystems, Warrington, UK; Foster City, CA). A genotyping assay based on the SNaPshot dNTP primer extension (PE Applied Biosystems) and restriction fragment length polymorphism (RFLP) methods were performed for genotyping of JAK3. Genotype study of JAK3 polymorphisms (-672 G>A and +227 G>A) were performed by the SNaPshot assay (primers shown in Appendix 2). Also, genotyping of +64 A>G polymorphisms were carried out by PCR-RFLP, which was done using a set of primers +64 A>G, 5'-CTGGGTG CAAAATTAGTTCCA-3'(sense), and 5'-CATCGCC AGCT CTTACCTAGC-3'(antisense) to generate a 231 bp fragment. Each PCR product (10 µL) was digested with 0.5 U of Msc1 according to manufacturer instructions at 37℃ for 8 hours. Following that, the digests were subjected to 2% agarose gel electrophoresis and stained with ethidium bromide to visualize allele-specific fragments.
3. Statistical analysis
Allele frequencies, genotype frequencies, and departures of the genotype distribution form Hardy-Weinberg equilibrium for each polymorphism were analyzed using the chi-square test or Fisher's exact test. Pairwise linkage disequilibrium (LD) for calculating D' and r2 was evaluated as described previously [15]. Linkage disequilibrium (|D'|) was calculated by the Haploview program ver. 3.2 (http://www.broad.mit.edu/mpg/haploview). Genotype-specific risks were estimated as odds ratios with associated 95% confidence intervals using unconditional logistic regression analysis ver. 8.02 (SAS Institute, Cary, NC) and they were adjusted for age and sex. p-value of <0.05 was considered statistically significant. p-values were also calculated for multiple testing using Bonferroni s inequality method.
Results
The demographics of the cases and controls enrolled in this study are shown in Table 1. We screened the promoter region of the JAK3 gene for polymorphisms in a small sample set of 24 lung cancer cases, and found 8 different JAK3 polymorphisms, including five novel polymorphisms. Among the 8 polymorphisms, 3 polymorphisms (-672 G>A, rs6512226; +64 A>G, rs7254346; and +227 G>A, rs7250423) were selected for large-scale genotyping, based on their frequencies (>25%), LD and haplotype tagging status (Table 2, Fig. 1). Genotype frequencies for case and controls were in Hardy-Weinberg equilibrium. Linkage disequilibrium coefficients (|D'|) between the polymorphisms were calculated using the Haploview program.
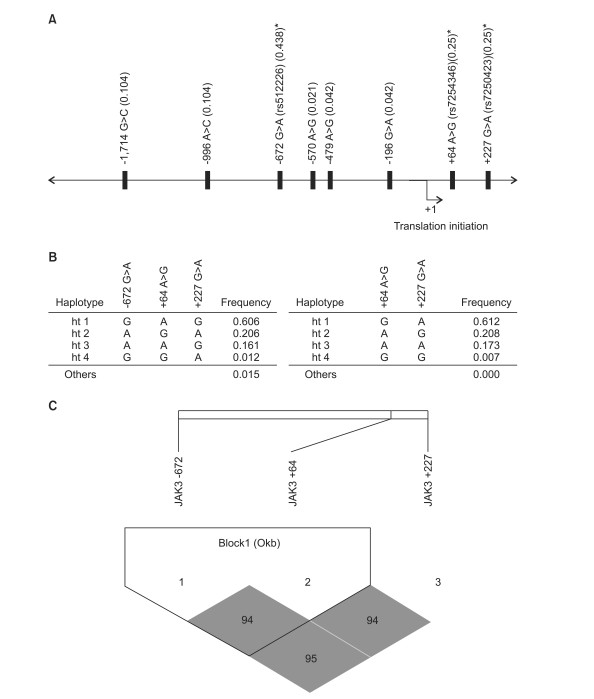
Map of polymorphisms, haplotypes, and linkage disequilibrium (LD) coefficients in janus tyrosine kinase3 (JAK3) gene. (A) The location of 8 polymorphisms in the JAK3 gene on chromosome 19p13.1. Asterisks indicated polymorphisms that were genotyped in a larger population. The frequencies of polymorphisms were based on sequencing data (n=24). The first base of the translation site was denoted as nucleotide +1. (B) Haplotypes of the promoter region in JAK3 gene. (C) Linkage disequilibrium coefficient (|D'|) among JAK3 gene. Two polymorphisms, including +64 A>G and +227 G>A were used for construction of haplotype. The LD between the polymorphisms was quantified using the Haploview program ver. 3.2.
Three polymorphisms were used for haplotype construction. The allelic frequencies of each polymorphism and haplotype were compared between the patients and controls using logistic regression models (Table 3). In genotype analyses, -672 G>A, +64 A>G, and +227 G>A were not associated risks of lung cancer in the overall and all subgroup analyses. The subsequent analysis showed that haplotype including 3 genotypes was not associated with an increased risk of lung cancer overall. However, further haplotype analysis revealed that haplotype 3 (+64A and +227A) was marginally associated with increased lung cancer risk in females, non-smokers and non-drinkers (Table 4). Other haplotypes including haplotype 1 (+64G and +227A), haplotype 2 (+64A and +227G), and haplotype 4 (+64G and +227G) remained not significant after stratifying by clinic pathological parameters (data not shown).
Discussion
In this study, we hypothesized that JAK3 polymorphisms were associated with lung cancer risk and that these polymorphisms may play a role as predictors of lung cancer. Here, we analyzed 3 polymorphisms of JAK3 promoter region by using genomic DNA from representatives of the Korean population. Our findings indicated no significant association between polymorphisms of the JAK3 gene and lung cancer risk. The small number of study subjects may have affected the lack of association.
Accumulating evidence indicated that dysregulation of the JAK-STAT signaling pathway caused cancers including lung cancer [10,16]. Studies have also shown that the JAKs interact with other well-known mitogenic pathways such as Raf/MEK signaling in the pathogenesis of malignancies [17,18]. Nevertheless, most JAK3 studies focused on leukemias, immune related diseases and lymphomas [11,19] and a relatively small number of studies were performed in solid cancer [16,20]. Recently, it has been suggested that the JAK-STAT pathway may be an effective target to control abnormal cell proliferation in lung cancer or pulmonary fibrosis through neuregulin-1 activation [21]. Despite their potential importance, polymorphisms of JAK3 in particular have not been examined. We therefore attempted to investigate a possible association between polymorphisms of JAK3 promoter and risk of lung cancer. Our data suggested that some haplotypes of JAK3 promoter were associated with an increased risk of lung cancer in the Korean population. This study is, to the best of our knowledge, the first report providing evidence for an association of the JAK3 promoter polymorphism with lung cancer risk.
Although cigarette smoking is considered one of the most important risk factors of lung cancer, previous studies have suggested genetic difference in epidemiologic characteristics such as non-smoking contributes to the pathogenesis of lung cancer [22,23]. Also, the associations between alcohol drinking and lung cancer risk have also been reported [11,24]. In our previous study, we showed that polymorphisms of the Her2 gene are associated with an increased susceptibility to lung cancer in females, non-smokers, and non-drinkers in the Korean population [15]. In the present study, haplotypes of the JAK3 gene increase the susceptibility to lung carcinogenesis on females, non-smokers, and non-drinkers. This suggests that genetic constitution of individuals is important in determining their susceptibility to lung cancer [25].
Conclusion
In this study, we identified 8 variations, including 5 novel polymorphisms, in the JAK3 promoter from 819 Korean subjects. Our data showed that haplotypes (-672 G>A and +64 A>G) are marginally associated with the increased risk of lung cancer, particularly in subgroups of the Korean population. However, we concluded that no significant association existed between these JAK3 polymorphisms and lung cancer risk in the Korean population.
Acknowledgments
We thank Dr. Jae Won Lee and Hyo Jung Lee for their assistance with the statistical analyses performed. This study was supported by the grant of the Korea Healthy 21 R&D Project, Ministry of Healthy & Welfare, Republic of Korea (A010250) and Seoul Research and Business Development Program (10574).
Notes
Conflict of interest relevant to this article was not reported.
Appendices
Appendix 1
Primer sequences for JAK3 variants screening
| SNP | PCR volume | Annealing temperature (℃) | Primer | |
|---|---|---|---|---|
| Forward | Reverse | |||
| -1,714 G>C | 10 μL PCR with Betaine | 60 | CAGGAAACAGCTATGACCGGCA TGACCACAGCTAAC |
TGTAAAACGACGGCCAG TTAATCTGGAGCCACAGG |
| -996 A>C | 10 μL PCR with Betaine | 60 | CAGGAAACAGCTATGACCCC CAAGTCTCTGCATTTG |
TGTAAAACGACGGCCAGTTGG TGATGCCTGTAATCC |
| -672 G>A | 10 μL PCR with Betaine | 60 | CAGGAAACAGCTATGACCTAA ACGAAGTCCCGCTCT |
TGTAAAACGACGGCCAG TAGACAGGCTGCTGGAGA |
| -570 A>G | 10 μL PCR with Betaine | 60 | CAGGAAACAGCTATGACC TAAACGAAGTCCCGCTCT |
TGTAAAACGACGGCCAGTA GACAGGCTGCTGGAGA |
| -479 A>G | 10 μL PCR with Betaine | 60 | CAGGAAACAGCTATGACCT AAACGAAGTCCCGCTCT |
GTAGACAGGCTGCTGGAGA TGTAAAACGACGGCCA |
| -196 G>A | 10 μL PCR with Betaine | 60 | CAGGAAACAGCTATGACCC CCAACTCACACATGCTAC |
TGTAAAACGACGGCC AGTATGCGCAATGACTCCTC |
| +64 A>G | 10 μL PCR with Betaine | 60 | CAGGAAACAGCTATGACC CCCAACTCACACATGCTAC |
TGTAAAACGACGGCCA GTATGCGCAATGACTCCTC |
| +227 G>A | 10 μL PCR with Betaine | 60 | CAGGAAACAGCTATGACCC CCAACTCACACATGCTAC |
TGTAAAACGACGGCC AGTATGCGCAATGACTCCTC |
JAK3, janus tyrosine kinase3; SNP, single nucleotide polymorphism; PCR, polymerase chain reaction.
Appendix 2
Genotyping primer sequences for JAK3 variants screening
| SNP | Annealing temperature (℃) | Strand | Primer | Addtives |
|---|---|---|---|---|
| -672 G>A | 60 | Forward | ATCACCAGGCCTGGCTAATTTTCCT | With betaine |
| +227 G>A | 55 | Reverse | GGATGCGAGTCTCGGCTCACGTCTG | - |
JAK3, janus tyrosine kinase3; SNP, single nucleotide polymorphism.



