AbstractPurposeMixed-lineage leukemia protein 4 (MLL4/KMT2D) is a histone methyltransferase, and its mutation has been reported to be associated with a poor prognosis in many cancers, including lung cancer. We investigated the function of MLL4 in lung carcinogenesis.
Materials and MethodsRNA sequencing (RNA-seq) in A549 cells transfected with control siRNA or MLL4 siRNA was performed. Also, we used EdU incorporation assay, colony formation assays, growth curve analysis, transwell invasion assays, immunohistochemical staining, and in vivo bioluminescence assay to investigate the function of MLL4 in lung carcinogenesis.
ResultsWe found that MLL4 expression was downregulated in non–small cell lung cancer (NSCLC) tissues compared to adjacent normal tissues and tended to decrease with disease stage progression. We analyzed the transcriptomes in control and MLL4-deficient cells using high-throughput RNA deep sequencing (RNA-seq) and identified a cohort of target genes, such as SOX2, ATF1, FOXP4, PIK3IP1, SIRT4, TENT5B, and LFNG, some of which are related to proliferation and metastasis. Our results showed that low expression of MLL4 promotes NSCLC cell proliferation and metastasis and is required for the maintenance of NSCLC stem cell properties.
IntroductionLung cancer is one of the most common malignant tumors worldwide. Lung cancer, which has a death rate of approximately 20%, is the leading cause of cancer-related deaths [1]. Generally, more than 80% of lung cancer is categorized as non–small cell lung cancer (NSCLC), and small cell lung cancer (SCLC) accounts for approximately 15% [2]. NSCLC can be further classified as adenocarcinoma, squamous cell carcinoma, and large cell carcinoma. Early detection and surgery offer the best chance for survival, and the 5-year survival rate is as high as 80%. However, most patients with lung cancer are diagnosed at an advanced stage with metastasis, and the 5-year survival rate is less than 10%. Over the past decade, low-dose computed tompgrahy has been the most commonly used screening method for lung cancer, improving early detection and reducing mortality [3]. However, low-dose computed tomography is far from satisfactory as a screening tool for clinical application because of its low specificity [3]. Therefore, there is an urgent need to identify novel potential biomarkers for lung cancer.
Post-translational histone modifications, which are a common epigenetic mechanism, are essential for establishing and maintaining the landscape of cell fate and identity [4]. Disruption of epigenetic modification genes, which are critical for proliferation, development, and differentiation, may lead to a loss of proliferative control, ultimately driving carcinogenesis [5]. Over the past several years, large-scale tumor DNA sequencing studies have found that the epigenetic regulator mixed-lineage leukemia protein 4 (MLL4/KMT2D) is one of the most frequently mutated genes in human cancer [6,7]. MLL4 is a major histone methyltransferase responsible for catalyzing H3K4me1 at transcription enhancers throughout the human genome [8]. MLL4 is a catalytic component of the mammalian complex of protein associated with Set1 (COMPASS) complex and is highly conserved throughout eukaryotes. This gene is homologous to the Trithorax-related gene in Drosophila. MLL4 protein sequences contain 5537aa and show 90% identity between humans and mice [9]. The MLL4 protein contains an enzymatically active C-terminal SET domain and two clusters of plant homeotic domains (PHDs) in the N-terminus region (three PHDs per cluster) [10]. MLL4 is enriched at enhancers of multiple genes and epigenetically regulates gene expression involved in multiple pathways, including the p53 pathway, cAMP-mediated signaling pathway, and cholestasis signaling pathway [11,12]. Interestingly, MLL4 performs opposite functions in different types of cancers. MLL4 can effectively inhibit the occurrence and metastasis of lymphomas, ovary cancer, and bladder cancer as a tumor suppressor [13–15]. While in prostate cancer, gastric cancer, breast cancer, pancreatic adenocarcinoma, and colorectal cancer, MLL4 is indispensable for the growth of tumor cells [6,11,12,16]. More recently, it has been reported that MLL4 could impede the tumorigenicity of human lung cancer cells by regulating multiple glycolytic genes [17].
In this study, we demonstrated that MLL4 functions as a tumor suppresser in NSCLC. We found that MLL4 has lower expression in NSCLC tissues than adjacent tissues and negatively correlated with NSCLC TNM grade. We revealed that MLL4 regulates a cohort of genes including PIK3IP1, which is a negative regulator of the phosphoinositide 3-kinase (PI3K)–AKT pathway and critically involved in cell proliferation and invasion. Our study revealed that MLL4 suppresses the tumor growth and metastasis by regulation of PI3K/AKT/SRY-related HMG-box 2 (SOX2) signaling in NSCLC cells.
Materials and Methods1. Cell culture and transfectionA549 and SK-MES-1 cell lines were purchased from the Chinese Academy of Medical Sciences, Shanghai, China. A549 cells were cultured in F12K medium (Gibco/Invitrogen, Gaithersburg, MD), and SK-MES-1 cells were cultured in MEM medium (Gibco/Invitrogen). All media were supplemented with 10% fetal bovine serum (FBS; Gibco/Invitrogen), 100 units/mL penicillin, and 100 mg/mL streptomycin (Gibco BRL). Cells were cultured at 37°C in a humidified incubator equilibrated with 5% CO2. Transfection was performed using Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. D-Luciferin Firefly was purchased from Perkin Elmer (Waltham, MA). PI3K inhibitor was purchased from Beyotime (Nantong, China). CRISPRa kit was purchased from OriGene (cat No. GA105405). The siRNAs were purchased from Sigma-Aldrich (St. Louis, MO). siRNA sequences are listed in S1 Table. PI3K inhibitor (ZSTK474), epidermal growth factor receptor (EGFR) inhibitor (gefitinib, osimertinib, erlotinib) was purchased from MCE MedChemExpress. All the experiments were performed in triplicate and repeated at least three times.
2. Lentiviral production and infectionRecombinant lentiviruses were constructed by Shanghai GenePharma (Shanghai, China). Concentrated viruses were used to infect 5×105 cells in a 60-mm dish with 8 μg/mL polybrene. Infected cells underwent sorting for target expression. shRNA sequences are listed in S1 Table.
3. Antibodies and reagentsThe antibodies used were anti-MLL4 (Sinalway Antibody, Baltimore, MD), anti-AKT, anti–p-AKT (Cell Signaling Technology, Danvers, MA), anti-Histone H3, anti-SOX2 (Abcam, Cambridge, MA), anti-MLL4, anti-PIK3IP1, anti-THBS1, anti-β-actin (Proteintech, Rosemont, IL), anti-MLL4 (Sigma-Aldrich), and anti-p53 (Santa Cruz Biotechnology, Santa Cruz, CA).
5. ImmunoprecipitationCellular extracts were harvested and incubated with the appropriate primary antibody or normal mouse/rabbit IgG at 4°C overnight. Samples were mixed with protein A/G magnetic beads for 2 hours at 4°C, and following a wash, the beads underwent sodium dodecyl sulfate polyacrylamide gel electrophoresis, followed by immunoblotting with a secondary antibody. Immunodetection was performed using enhanced chemiluminescence with an enhanced chemiluminesence system (cat. No. WBKLS0500, Millipore, Billerica, MA) according to the manufacturer’s instructions.
7. EdU incorporation assayBriefly, cells were cultured with 5-ethynyl-2′-deoxyuridine (EdU) for 2 hours before detection. The proliferation rate of the cells was then evaluated using a Cell-Light EdU Cell Proliferation Detection kit (RiboBio, Guangzhou, China) following the manufacturer’s instructions.
8. Cell invasion assayBriefly, 2×104 cells were seeded into the upper chamber in a volume of 300 μL. Cells on the upper side of the membrane were removed by cotton swabs and those on the other side were stained and counted.
9. In vitro wound-healing assayBriefly, cells were grown to confluence and wounds were made using sterile pipette tips. Cells were washed with phosphate buffered saline (PBS) and incubated in the fresh medium without FBS. After 36 hours of incubation at 37°C, the cells were imaged. Data shown are the mean±standard deviation (SD) for n=6 wells per group.
10. CRISPR activation transfection in cellsCRISPR activation (CRISPRa) is one type of CRISPR tool that uses the enzymatically deficient Cas9, a mutation of Cas9 without endonuclease activity, CRISPRa system can be used to activate endogenous gene expression. Approximately 18–24 hours before transfection, 3×105 adherent cells were plated in 2 mL culture media into each well of a 6-well plate. One microgram of MLL4 gRNA vector (or Scramble gRNA) and 0.3 μg enhancer vector were diluted in 250 μL of Opti-MEM and gently vortexed. And 5 μL of Turbofectin was added to the diluted DNA (not the reverse order) and pipetted gently to mix completely. The mixture was incubated for 15 minutes at room temperature. Then, the mixture was added drop-wise onto the plated cells. Forty-eight hours post-transfection, gene expression can be measured with quantitative real-time PCR (qPCR).
11. Sphere formation assayA549 or SK-MES-1 cells were inoculated into a 1:1 mixture of DMEM/F12 medium (Gibco/Invitrogen) without serum containing B27 supplement (50×, Invitrogen), 0.4% BSA, 4 mg/mL insulin (Invitrogen), human recombinant epidermal growth factor (20 ng/mL), and basic fibroblast growth factor (10 ng/mL) at a density of 1,000 cells/well in ultra-low attachment 6-well plates (Corning, Inc., Corning, NY). The size and number of spheroids were analyzed under an optical microscope. Spheroids more than 30 μm in size are the criteria for sphere formation.
12. Tissue specimens and immunochemistryAll human tissue samples were collected using protocols approved by the Ethics Committee of Lishui Hospital, and informed consent was obtained from all patients. Samples were frozen in liquid nitrogen immediately after surgical removal and maintained at −80°C until analysis. Samples were fixed in 4% paraformaldehyde (Sigma-Aldrich) at 4°C overnight, embedded in paraffin, sectioned at 8 μm onto Superfrost-Plus Slides, which were then processed per standard protocols using 3,3’-diaminobenzidine staining and monitored microscopically.
13. Tumor xenografts4×106 A549 cells in 100 μL of PBS were injected subcutaneously into the 6–8-week-old BALB/c nude mice (Vital River, Beijing, China). Five animals per group were used in each experiment. Tumors were measured every 3 days using a vernier caliper, and the volume was calculated according to the following formula: π/6×length×width2. All studies were approved by the Animal Care Committee of Wenzhou Medical University.
14. In vivo metastasisA549 cells that had been transfected to stably express firefly luciferase (Xenogen, Alameda, CA) were infected with lentiviruses carrying MLL4 shRNA or scrambled control shRNA. These cells were injected into the lateral tail vein (5×106 cells) of 6-week-old male nonobese diabetic/severe combined immunodeficiency (NOD SCID) mice. For bioluminescence imaging, mice were injected with 200 mg/g of D-luciferin in PBS abdominally. At 10 minutes after injection, mice were anesthetized and bioluminescence was imaged with a charge-coupled device camera (IVIS; Xenogen).
Results1. The expression of MLL4 is downregulated in NSCLCTo determine the expression of MLL4 in lung cancer, we analyzed the expression level of MLL4 in three published clinical datasets (GSE33356, GSE19188, and GSE18442) from the Gene Expression Omnibus (GEO) database (Fig. 1A). The results revealed that MLL4 expression was downregulated in lung cancer tissues compared with normal lung tissues. In addition, we found the mutation frequency of MLL4 was high in different lung cancer type databases (Fig. 1B). To confirm the result, we collected 16 pairs of NSCLC and adjacent normal tissues and found that the mRNA level of MLL4 was downregulated in the tumor tissues (Fig. 1C). Immunohistochemical analysis also revealed lower expression in the tumor tissues than the adjacent normal tissues (Fig. 1D). Then, we explored the protein expression level of MLL4 in 35 pairs of NSCLC and adjacent normal tissues with TNM clinical classification using immunohistochemistry staining (Fig. 1E). The results indicated a decreased expression level of MLL4 in the tumor tissues compared to the adjacent normal tissues, and its expression tended to be negatively correlated with disease stage progression (Fig. 1F and G).
2. Transcriptome analysis of MLL4-regulated genes and signaling pathways in NSCLCTo determine how MLL4 regulates non–small cell lung carcinogenesis, we investigated the transcriptomes in the control and MLL4 siRNA A549 cells using a high-throughput RNA deep sequencing approach (RNA-seq). Total RNA was extracted from A549 cells transfected with control siRNA or siRNA-targeting MLL4, and three independent samples and controls were assessed. Whole-transcriptome clustering analysis revealed that the expression levels of 1,042 genes were significantly changed upon MLL4 knockdown (Fig. 2A). A total of 515 genes showed up-regulated expression, whereas 527 genes showed downregulated expression in the A549 cells treated with MLL4 siRNA compared with samples treated with control siRNA. Gene set enrichment analyses showed significant enrichment in transforming growth factor β (TGF-β) signaling pathways after MLL4 knockdown (Fig. 2B). Furthermore, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were conducted based on both genes with up-regulated and downregulated expression, revealing the vital 12 enriched pathways. These signaling pathways include the PI3K-AKT, tumor necrosis factor, advanced glycation end-products–receptor for advanced glycation end products, TGF-β, and interleukin-17 signaling pathways (Fig. 2C). To validate the RNA-seq analysis, we chose 19 representative genes (Fig. 2D) and validated their expression in A549 and SK-MES-1 cells using qPCR. The results indicated that the mRNA levels of ATF1, FOXP4, KDM2A, SOX2, SOX21, PABL6, SMAD9, BMP6 and THBS1 increased, while the RAB37, RASSF4, PACR3, PIK3IP1, SIRT4, TENT5B, LFNG, RBL1, BATF2 and CXXC5 levels decreased upon the knockdown of MLL4 (Fig. 2E and F). Western blot demonstrated that PIK3IP1 expression was downregulated following MLL4 knockdown, whereas the P-AKT, SOX2, and THBS1 levels were up-regulated and the expression of AKT was unchanged (Fig. 2G). To determine whether MLL4 could affect the global level of H3K4me1 in NSCLC cells, cells were transfected with siRNA specifically targeting MLL4. The results showed that knockdown of MLL4 led to a decreased expression of global H3K4me1 level in A549 and SK-MES-1 cells (Fig. 2H). Since MLL4 regulates gene expression mainly through binding and methylation of enhancer regions, we also performed ChIP assay using antibodies against MLL4 or control IgG. The results revealed that MLL4 does not occupy at the classical enhancers (SSR1 and SSR2) [20] and promoters (SOX2-promoter) region of SOX2 gene (S4A Fig.). In order to determine whether the regulation of SOX2 by MLL4 was dependent on PIK3IP1. Using western blotting, we found that the increased expression of SOX2 with MLL4 knockdown, which could be restored by combining with PIK3IP1 overexpression (Fig. 2I). In addition, we also detected the expression of SOX2 in the MLL4 knockdown A549 and SK-MES-1 cells by treating the PI3K inhibitor (ZSTK474) (Fig. 2J). The results showed that the expression of SOX2 was significantly decreased after treatment with PI3K inhibitor in both control siRNA and MLL4 siRNA cells. These results suggested that MLL4 regulates the expression of SOX2 in PIK3IP1-dependent manner and MLL4 may regulate the progression of NSCLC by regulating the PI3K/AKT/SOX2 axis.
3. Stable inhibition of MLL4 promotes the proliferation of NSCLC cellsTo investigate the function of MLL4 in NSCLC, we evaluated cell proliferation in the NSCLC cells. We used CRISPRa to activate the expression of MLL4 and used the lentivirus-mediated stable knockdown efficiencies of MLL4 by two MLL4-specific shRNA (#1 and #2). The results of EdU staining demonstrated that the percentage of EdU-positive cells was significantly decreased in MLL4 overexpression A459 cells and increased in the MLL4-silenced A549 cells. Furthermore, in agreement with the functional link between MLL4 and PIK3IP1 described previously, the increase in EdU-positive cells resulting from MLL4 knockdown was partially reversed by overexpression of PIK3IP1 (Fig. 3A). In addition, consistent results were obtained in SK-MES-1 cells (Fig. 3B). Colony formation assays further showed that shRNA knockdown of MLL4 increased the colony formation ability, while could be restored by combining with PIK3IP1 overexpression in A549 cells and SK-MES-1 cells (Fig. 3C and D). We confirmed the efficiency of lentivirus-mediated shRNAs targeting MLL4 and CRISPRa-MLL4 gRNA using reverse transcription polymerase chain reaction (RT-PCR), and the expression level of PIK3IP1 overexpression was detected by western blot analysis (Fig. 3E). Moreover, EdU staining assays (Fig. 4A and B) and colony formation assays (Fig. 4C and D) also showed that knockdown of PIK3IP1 or overexpression of SOX2 mimics the phenotype of MLL4 knockdown in A549 cells and SK-MES-1 cells. The western blots shown in Fig. 4E further verified the efficacies of the shRNAs and plasmids used in these experiments. In addition, growth curve assays also showed that MLL4 knockdown significantly increased the growth of A549 cells and SK-MES-1 cells, which could be partially alleviated by overexpression of PIK3IP1 (Fig. 5A and B).
4. Knockdown of MLL4 promotes the metastasis of NSCLC cellsNext, we investigated the role of MLL4 in tumor migration and invasion. For this purpose, CRISPRa was used to activate the expression of MLL4 and MLL4 knockdown by MLL4-specific shRNA in A549 and SK-MES-1 cells. In the wound-healing assays, the amount of open distance remaining after 36 hours of migration was different compared to that of the control. The overexpression of MLL4 resulted in a significant delay in wound closure in A549 and SK-MES-1 cells, whereas MLL4 knockdown was associated with an increased migration rate, which could be partially rescued by overexpression of PIK3IP1 (Fig. 5C and D). Furthermore, the results from transwell invasion assays in two NSCLC cell lines (A549 and SK-MES-1) showed a significantly decreased invasiveness caused by MLL4-CRISPRa. Moreover, we also found that a significant increase in invasiveness was associated with MLL4 knockdown, which could be restored by combining with PIK3IP1 overexpression (Fig. 5E and F). RT-PCR and western blot shown in Fig. 5G further verified the efficacies of the indicated shRNAs, CRISPRa-MLL4 gRNA, and plasmids used in these experiments. Meanwhile, PIK3IP1 knockdown and SOX2 overexpression showed increased migration rate (Fig. 6A and B) and invasiveness (Fig. 6C and D) compared to the control groups, consistent with the phenotype of MLL4 knockdown. The western blots shown in Fig. 6E further verified the efficacies of the shRNAs and plasmids used in these experiments. These data are consistent with a role of MLL4 in suppressing carcinogenesis and support the observation that PIK3IP1 is a downstream target of MLL4.
5. MLL4 knockdown has a positive effect on sphere formation of NSCLC cellsSphere formation assay is an in vitro method commonly used to identify CSCs and study their properties. Since SOX2, the target gene regulated by MLL4, has an important role in maintaining stem cell properties and functions, we evaluated the role of MLL4 in the sphere forming ability of A549 and SK-MES-1 cells. The results showed that knockdown of MLL4 increased the number and size of the spheres of A549 (Fig. 7A) and SK-MES-1 cells (Fig. 7B) compared to the control cells in low-attachment plates. Moreover, the increased size and number of spheres with MLL4-targeted shRNA was markedly blocked by co-transfected with PIK3IP1. The expression level of MLL4 and PIK3IP1 was further verified through RT-qPCR and Western blot (Fig. 7C). In addition, knockdown of SOX2 decreased the number and size of the spheres, and PIK3IP1 knockdown and SOX2 overexpression showed increased the number and size of the spheres in A549 (Fig. 8A) and SK-MES-1 cells (Fig. 8B). The western blots shown in Fig. 8C further verified the efficacies of the shRNAs and plasmids used in these experiments. Based on these results, low expression of MLL4 may be required for the maintenance of NSCLC stem cell properties, and it does so, at least in part, through regulating the PI3K/AKT/SOX2 axis.
6. Stable inhibition of MLL4 promotes tumor growth and metastasis in vivoFurthermore, we investigated the role of MLL4 in tumor development and progression in vivo by subcutaneously implanting A549 cells that had been engineered to stably express MLL4 shRNA or control scrambled shRNA in athymic BALB/c mice. Growth of the implanted tumors was monitored by measuring the tumor sizes every 3 days over a period of 4 weeks. The results showed that tumor growth was substantially increased upon MLL4 knockdown (Fig. 9A). The average tumor volume/weight of mice in the MLL4 interference group was 2.59/1.93 times that of the control group. To further explore the role of MLL4 in NSCLC progression. We investigated the role of MLL4 in NSCLC metastasis in vivo. A549 cells that had been engineered to stably express firefly luciferase (A549-Luc) were infected with lentiviruses carrying shRNA against MLL4 and then injected intravenously through the tail vein of 6-week-old male NOD SCID mice. Lung metastasis was quantified using bioluminescence imaging after 4 weeks. The results showed that MLL4 knockdown led to a dramatic increase in lung metastasis of the A549-Luc tumors. The metastases to the lungs were verified by bioluminescence imaging and histological staining (Fig. 9B). In order to further extend our observation results to a clinic-pathologically relevant context, we analyzed the expression of MLL4, PIK3IP1, and SOX2 with clinical behavior in lung cancer patients. Kaplan-Meier survival analysis (http://kmplot.com/analysis/) shows that the overexpression of MLL4/PIK3IP1 or low expression of SOX2 was associated with improved overall survival in the lung cancer patients (Fig. 9C). In addition, a reanalysis of the data sourced from published clinical datasets such as GSE108492, GSE115457, GSE8894, and GSE3141 indicated that the expression of MLL4 is significantly positively correlated with PIK3IP1 expression and negatively correlated with SOX2 expression, supporting our finding that PIK3IP1 was transcriptionally regulated by MLL4 (Fig. 9D). Taken together, these findings indicated that low expression of MLL4 promotes NSCLC cell tumorigenesis in vivo.
DiscussionMLL4 is a histone methyltransferase, and mutations in this gene have been reported to associate with a poor prognosis in many cancers, including non-Hodgkin lymphoma, medulloblastoma, prostate cancer, renal cancer, bladder cancer, and lung cancer. The MLL4 mutation rate was 17.5% and associated with reduced survival in NSCLC [7]. Here, in our study, we found that the expression of MLL4 was downregulated in lung cancer by conducting an analysis of data from the GEO database. And mutation frequency of MLL4 was high in different lung cancer type databases. We demonstrated that MLL4 showed lower expression in NSCLC tissues than adjacent normal tissues, and the expression tended to decrease with disease stage progression. Through a series of cell experiments, we revealed that downregulated MLL4 expression accelerated NSCLC cell proliferation and metastasis. We found that knockdown of MLL4 increased the expression of a set of genes including ATF1, FOXP4, KDM2A, SOX2, SOX21, PABL6, SMAD9, BMP6, and THBS1 and decreased the expression of RAB37, RASSF4, PACR3, PIK3IP1, SIRT4, TENT5B, LFNG, RBL1, BATF2, and CXXC5. Some of the target genes are known to be critically involved in tumorigenesis and metastasis.
The PI3K/AKT signaling pathway plays a crucial role in regulating cell survival, growth, proliferation, altered adhesion, angiogenesis, transcription, translation, and metabolism [21]. This pathway is critical for human normal physiology, and its dysfunction could cause various human cancers [22]. It has been reported that the abnormal regulation of the PI3K/AKT pathway causes activation of other downstream signaling pathways linked with tumorigenesis; therefore, it is an important therapeutic target for discovery of potential antitumor agents [21]. In a pancancer analysis, the most common aberrations of this pathway were mutations and/or genomic amplification of AKT, loss of PTEN, and somatic activating mutations in PIK3CA [23]. Therefore, tumor growth and metastasis can be inhibited by suppressing the expression of each molecule in the pathway and blocking signaling, leading to an antitumor effect. In our study, we found that knockdown of MLL4 decreased the expression of PIK3IP1, which is a negative regulator of PI3K, and increased the expression of phosphorylated AKT. MLL4 may regulate the expression of PIK3IP1 at the transcriptional level and then influence the PI3K/AKT signaling pathway.
SOX2 belongs to a large family of the SOX transcription factors. It is well known for its critical role in maintaining an undifferentiated status for the pluripotency of ES cells or induced pluripotent stem (iPS) cells [24]. Recently, aberrant expression of SOX2 has been demonstrated in various types of human cancers, including lung cancer [25]. Overexpression of SOX2 has been described in all types of lung cancer tissues, including squamous cell carcinoma, adenocarcinoma, and SCLC tissues [26]. A combination of SOX2 overexpression and mammalian target of rapamycin pathway activation is one of the characteristics of lung squamous cell carcinoma tumorigenesis. Importantly, high expression of SOX2 plays a key role in conferring stem cell-like phenotypes in more than a dozen tumors [25]. In addition, SOX2 has been recognized as a novel target of EGFR-Src-AKT signaling in NSCLCs that regulates self-renewal of stem-like cells, indicating that the functions of SOX2 in the CSCs are a major determinant in EGFR-targeted therapy [26,27]. In our study, we also found that knockdown of MLL4 significantly increased the expression of SOX2 and sphere forming ability of lung cancer cells, indicating that a low expression level of MLL4 was important for maintenance of lung cancer cell stem cell properties. Interestingly, there were various inter-dependencies between PI3K/AKT signaling and SOX2 from different tumor cell, stem cell, and healthy cell types [28]. On the one hand, PI3K/AKT signaling–induced phosphorylation sustained nuclear SOX2 expression, accelerating tumor cell dissemination; on the other hand, SOX2 increased PI3KCA gene expression and then activated AKT signaling in glioblastoma [29]. SOX2 and KLF4 together drive PIK3CA gene expression, enhancing PI3K/AKT activity and tumorigenesis, and in turn, overexpression of PIK3CA rescues growth inhibition caused by deletion of SOX2 in nasopharyngeal carcinoma [30]. It is well known that iPS cell induction involves artificial overexpression of exogenously introduced SOX2, but SOX2 overexpression coincides with elevated AKT activity during reprogramming [31]. Thus, the PI3K/AKT/SOX2 signaling axis is a conserved module in cancer, stemness and reprograming. In our study, we found that knockdown of MLL4 downregulated the expression of PIK3IP1 and up-regulated the expression of phosphorylated AKT and SOX2. Therefore, downregulation of MLL4 influenced PI3K/AKT/SOX2 signaling, accelerating tumorigenesis. In addition, we found that MLL4 does not regulate the expression of SOX2 by directly binding to the promoter or classical enhancers (SSR1 and SSR2) of SOX2. And MLL4 regulates its expression mainly through PIK3IP1. To further determine the precise location of the enhancer region of the MLL4 target genes, MLL4 ChIP-seq assays should be performed in future studies. Whether MLL4 can regulate gene expression by methylating promoter regions is still unknown. In addition, we will further study the mechanism of how MLL4 regulates PIK3IP1 in NSCLC cells.
As EGFR is an important regulator of the PI3K/AKT signaling pathway, we treated lung cancer cells with the EGFR tyrosine kinase inhibitor (TKI) (gefitinib, osimertinib, and erlotinib) to check the expression of MLL4, and found that the expression of MLL4 was slightly decreased in EGFR TKI group compared to control, but there was no statistically significant. (S4B Fig.). Besides, Egolf et al. [32] reported that MLL4 could interact with p53, which is also important tumor suppressor in lung cancer. We performed immunocoprecipitation using cell lysates from human lung cancer cells. The results showed that p53 could co-immunoprecipitated with MLL4 (S4C Fig.). In addition, our RNA-seq analysis shown that p53 was not regulated by MLL4. To detect whether MLL4 and p53 could be regulated by each other, we texted the expression levels of MLL4 and p53 in response to p53 or MLL4 knockdown. The results showed that downregulation of MLL4 could not affect the expression of p53 at mRNA levels. Similarly, knockdown of p53 could not affect the MLL4 expression either (S4D Fig.). These results indicated that MLL4 could interact with p53, but could not regulate the expression of p53.
In summary, our data indicated that MLL4 regulates the PI3K/AKT/SOX2 signaling pathway, promoting NSCLC tumorigenesis and progression by regulating the expression of PIK3IP1 and SOX2. Furthermore, our findings support the pursuit of MLL4 as a potential prognostic indicator and/or target for the treatment of NSCLC.
Electronic Supplementary MaterialSupplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
NotesEthical Statement The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Lishui Hospital (protocol code 2019(64) and 2019/8/1). The animal study protocol was approved by the Animal Care Committee of Wenzhou Medical University (protocol code 18381 and 2019/7/29). Author Contributions Conceived and designed the analysis: Zhao Z, Ji J. Collected the data: Xu M. Contributed data or analysis tools: Yang Y, Qiu R, Xu Z, Zhao S, Zhang D, Wang H. Performed the analysis: Yang Y, Qiu R, Xu Z, Meng M, Kong C, Wang H. Wrote the paper: Yang Y, Qiu R. Methodology, review and editing, funding: Yang Y. Review and editing, funding: Qiu R. Resources: Weng Q, Song J. AcknowledgmentsThis work was supported by a grant (2020KY1085 to R.Q.) from the Medical and Health Science and Technology Program of Zhejiang Province, a grant (LY21H160010 to Y.Y.) from the Basic Public Welfare Research Program of Zhejiang Province, and a grant (2020ZDYF10 to R.Q.) from the Key R&D Program of Lishui City.
Fig. 1Mixed-lineage leukemia protein 4 (MLL4) expression is downregulated in non–small cell lung cancer (NSCLC) tissue. (A) Analysis of public datasets (GSE33356, GSE19188, and GSE18442) for the expression of MLL4 by two-tailed unpaired t test. (B) MLL4 alteration frequency from five lung cancer data sets, data analysis was performed on the ciBioportal platform. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MSKCC, Memorial Sloan Kettering Cancer Center; SCLC, small cell lung cancer; TCGA, The Cancer Genome Atlas. (C) Real-time polymerase chain reaction (PCR) of MLL4 in 16 pairs of NSCLC and adjacent normal tissues. The expression of MLL4 was measured by quantitative real-time PCR. In addition, mRNA levels were normalized to that of β-actin. Each bar represents the mean±standard deviation for triplicate experiments. (D) Immunohistochemical staining of MLL4 in 16 pairs of NSCLC and adjacent normal tissues. For each group, representative photos of two specimens are shown. (E) H&E staining and immunohistochemical staining of MLL4 in 35 pairs of NSCLC and adjacent normal tissues with TNM clinical classification (adjacent, stage 0, I, II, III, and IV). For each stage, representative photos of two specimens are shown. (F, G) The positively stained nuclei (%) in 35 paired samples were analyzed by two-tailed unpaired t test (*p < 0.05, **p < 0.01, ***p < 0.001). 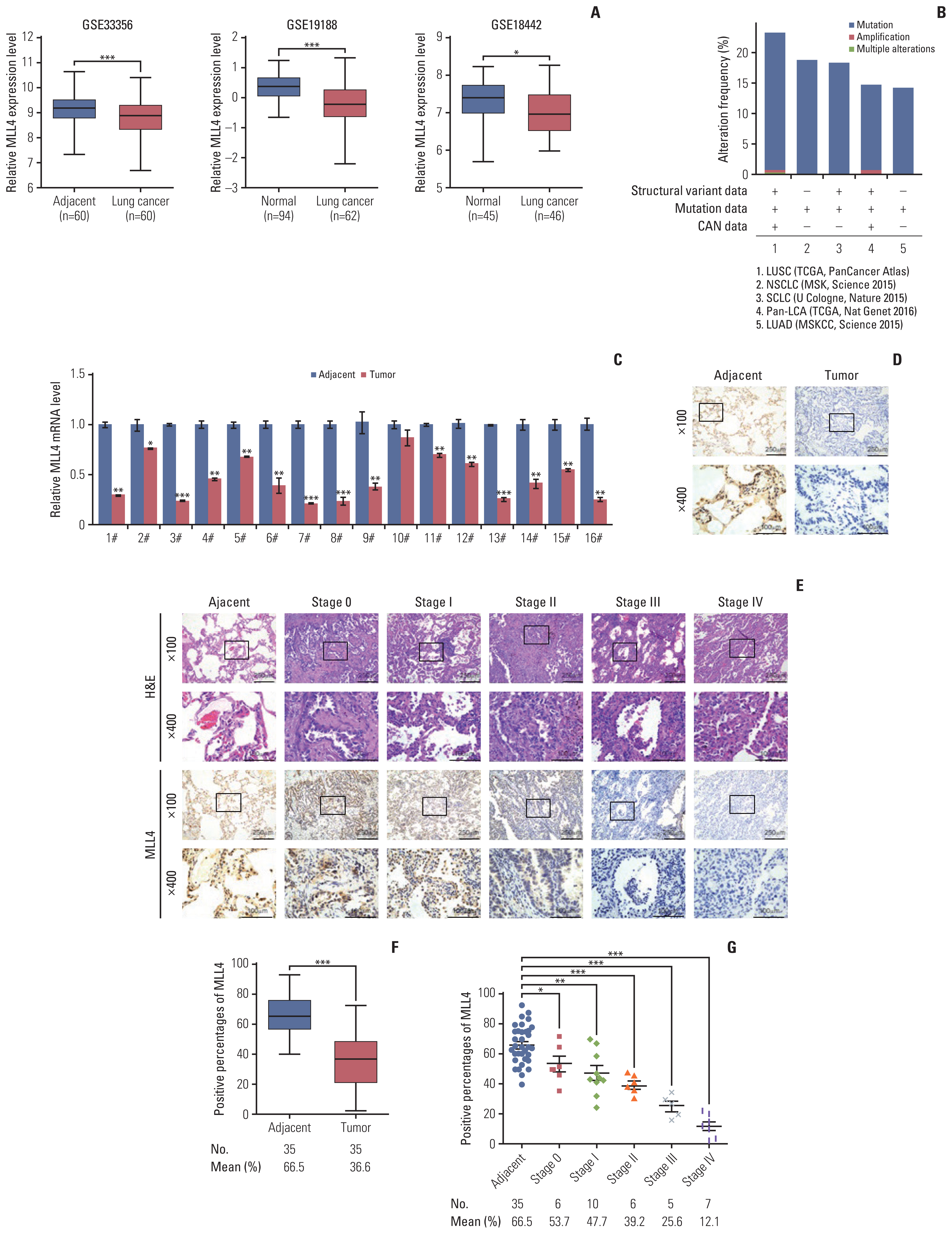
Fig. 2Genome-wide identification of transcription targets for mixed-lineage leukemia protein 4 (MLL4). (A) A549 cells were transfected with control siRNA and MLL4 siRNA followed by RNA extraction and deep sequencing. (B) Gene set enrichment analysis (GSEA) results showing enriched gene signatures related to the transforming growth factor β (TGF-β) signaling pathway in A549 cells infected with control siRNA or MLL4 siRNA. (C) MLL4-regulated genes were grouped and statistically analyzed according to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. (D) The heatmap presented the effect of MLL4 knockdown on the expression of genes. (E, F) A549 cells or SK-MES-1 cells were transfected with the indicated siRNAs, followed by RNA extraction and quantitative reverse transcription PCR analysis of the expression of the indicated genes. The mRNA levels were normalized to those of β-actin. *p < 0.05, **p < 0.01, and ***p < 0.001 (two-tailed t test). (G) A549 cells or SK-MES-1 cells were transfected with the indicated siRNAs, followed by western blot analysis of the expression of the indicated genes. (H) A549 cells or SK-MES-1 cells were transfected with MLL4 siRNA #2, and the protein levels of H3K4me1 were measured. H3 served as loading control for the western blot. (I) A549 cells transfected with indicated siRNAs and plasmid and Western blot analysis of indicated target genes. The knockdown efficiencies of MLL4 were confirmed by quantitative real-time PCR. (J) Control siRNA or MLL4 siRNA transfected A549 and SK-MES-1 cells were treated in the presence or absence of PI3K inhibitor (ZSTK474) at 10 μm concentration for 24 hours. The p-AKT and SOX2 levels are shown. β-Actin was used as a loading control. AGE-RAGE, advanced glycation end-products–receptor for advanced glycation end products; ECM, extracellular matrix; IL-17, interleukin 17; PI3K, phosphoinositide 3-kinase; TGF, transforming growth factor; TNF, tumor necrosis factor. 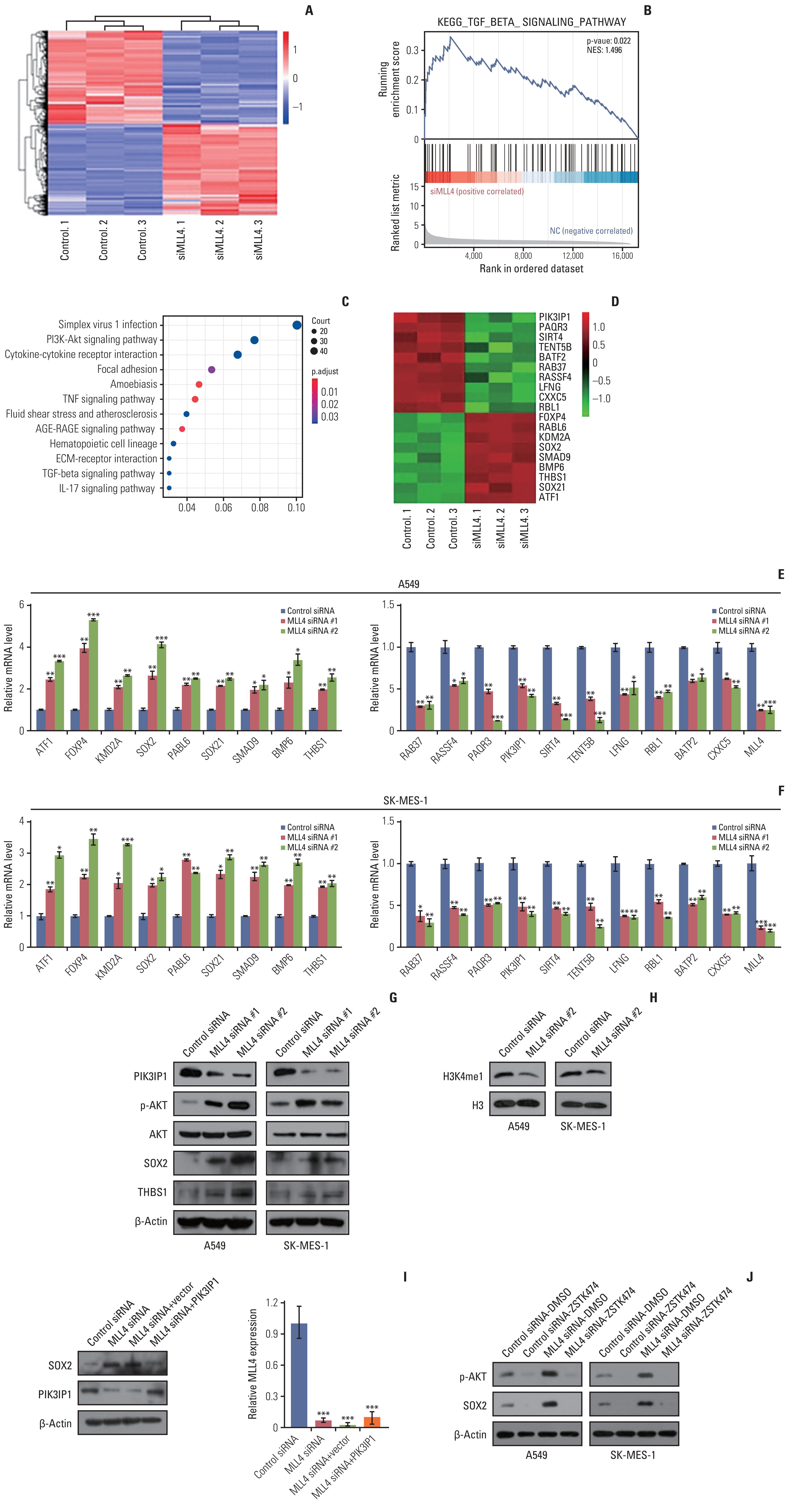
Fig. 3Stable inhibition of mixed-lineage leukemia protein 4 (MLL4) promotes the proliferation in non–small cell lung cancer cells. (A, B) A549 cells and SK-MES-1 cells were transfected with CRISPR activation (CRISPRa)-Scramble or CRISPRa-MLL4 gRNA or the indicated specific shRNAs or/and expression constructs for EdU assays. (C, D) A549 cells and SK-MES-1 cells stably infected with CRISPRa-Scramble or CRISPRa-MLL4 gRNA or the indicated specific shRNAs or/and expression constructs in culture media for 10 days prior to being stained with crystal violet. Representative photos and statistical analyses are shown. (E) The activation and knockdown efficiencies of MLL4 were confirmed by quantitative real-time PCR. The overexpression of PIK3IP1 was confirmed by western blot analysis. Each bar represents the mean±standard deviation for triplicate measurements. *p < 0.05 and **p < 0.01 (two-tailed t test). 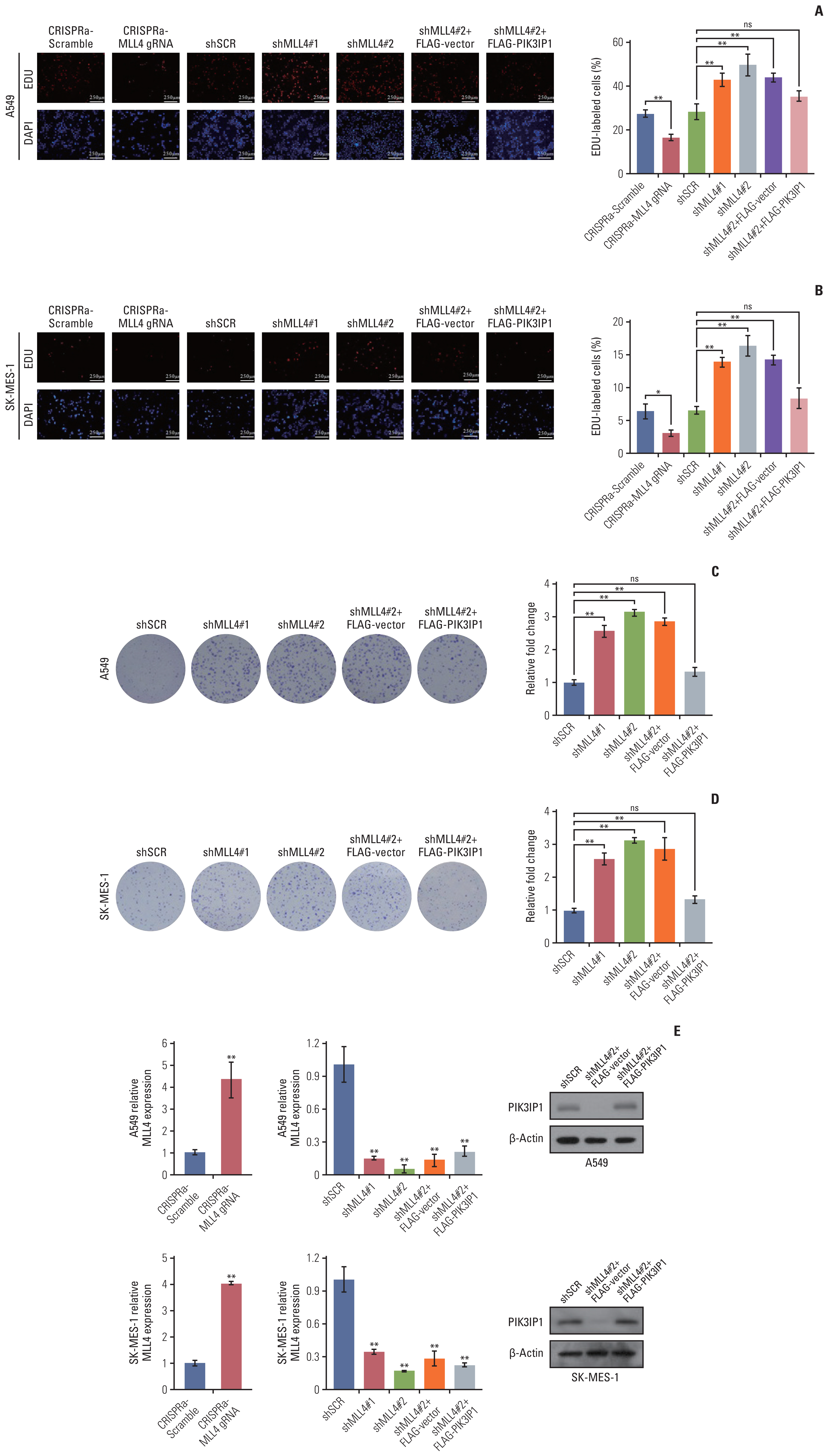
Fig. 4The effect of PIK3IP1 and SOX2 on non–small cell lung cancer cells proliferation. (A, B) A549 cells and SK-MES-1 cells were transfected with the indicated specific shRNAs or/and expression constructs for EdU assays. (C, D) A549 cells and SK-MES-1 cells stably infected with the indicated specific shRNAs or/and expression constructs in culture media for 10 days prior to being stained with crystal violet. (E) Western blot analysis was used to determine the protein expression in these cells using antibodies against the indicated proteins. Each bar represents the mean±standard deviation for triplicate measurements. **p < 0.01 (two-tailed t test). 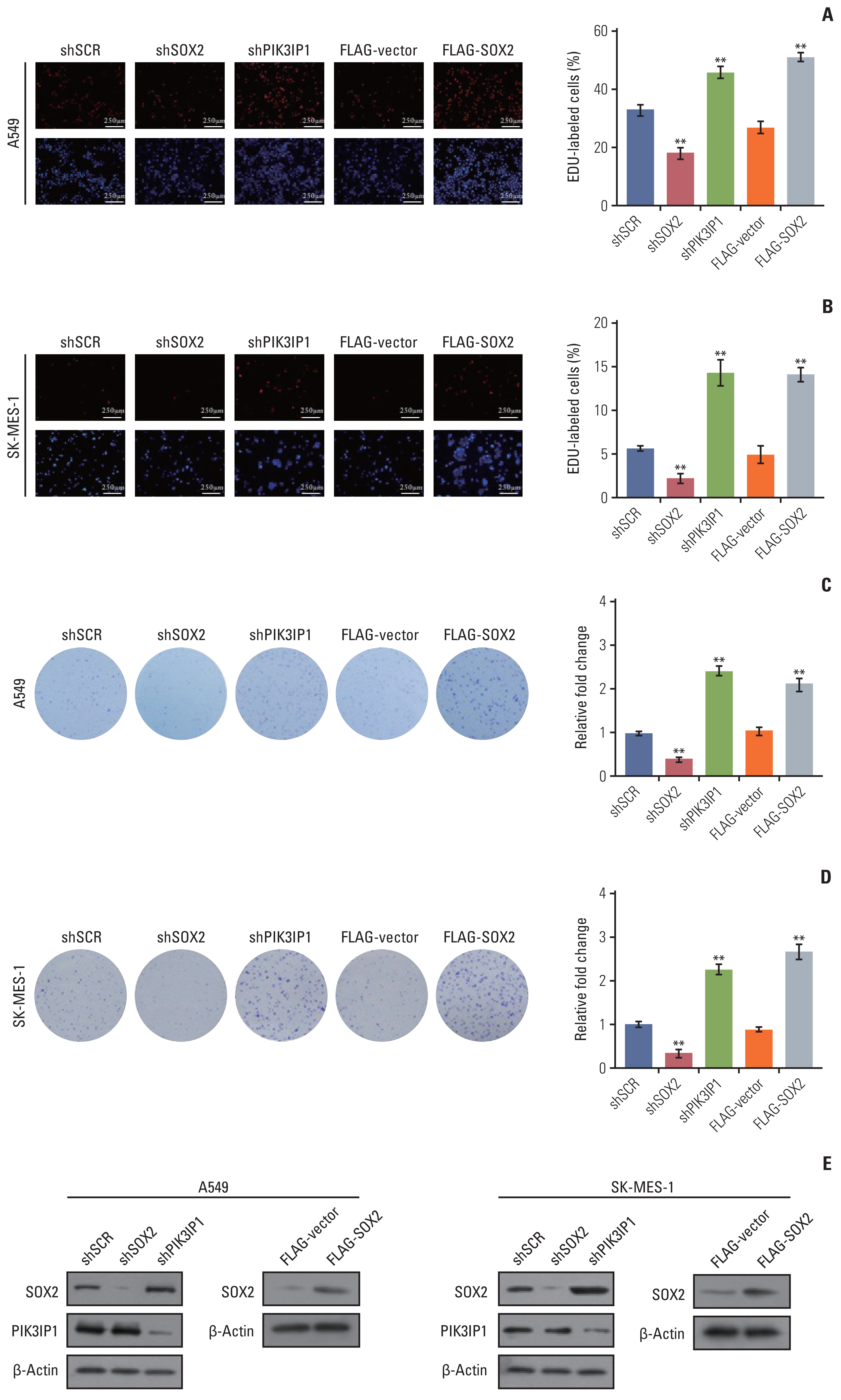
Fig. 5Knockdown of mixed-lineage leukemia protein 4 (MLL4) promotes the invasion and migration of non–small cell lung cancer cells. (A, B) A549 and SK-MES-1 cells were transfected with the indicated specific shRNAs or/and expression constructs subjected to growth curve analysis by counting the numbers of living cells. (C, D) A549 cells and SK-MES-1 cells were transfected with Scramble or MLL4 gRNA or the indicated specific shRNAs or/and expression constructs. The migration of cells towards the wound was photographed under a light microscope. Representative photos and statistical analyses are shown. (E, F) Transwell invasion assays of A549 and SK-MES-1 cells following transfection with CRISPR activation (CRISPRa)-Scramble or CRISPRa-MLL4 gRNA or the indicated specific shRNAs and/or expression constructs. **p < 0.01 and ***p < 0.001 (two-tailed t test). (G) The activation and knockdown efficiencies of MLL4 were confirmed by quantitative real-time polymerase chain reaction. The overexpression of PIK3IP1 was confirmed by western blot analysis. 
Fig. 6The effect of PIK3IP1 and SOX2 on non–small cell lung cancer cells migration and invasion. (A, B) A549 cells and SK-MES-1 cells were transfected with the indicated specific shRNAs or/and expression constructs. The migration of cells towards the wound was photographed under a light microscope. (C, D) Transwell invasion assays of A549 and SK-MES-1 cells following transfection with the indicated specific shRNAs or/and expression constructs. The invaded cells were stained and counted. (E) Western blot analysis was used to determine the protein expression in these cells using antibodies against the indicated proteins. Each bar represents the mean±standard deviation for triplicate measurements. *p < 0.05, **p < 0.01, and ***p < 0.001 (two-tailed t test). 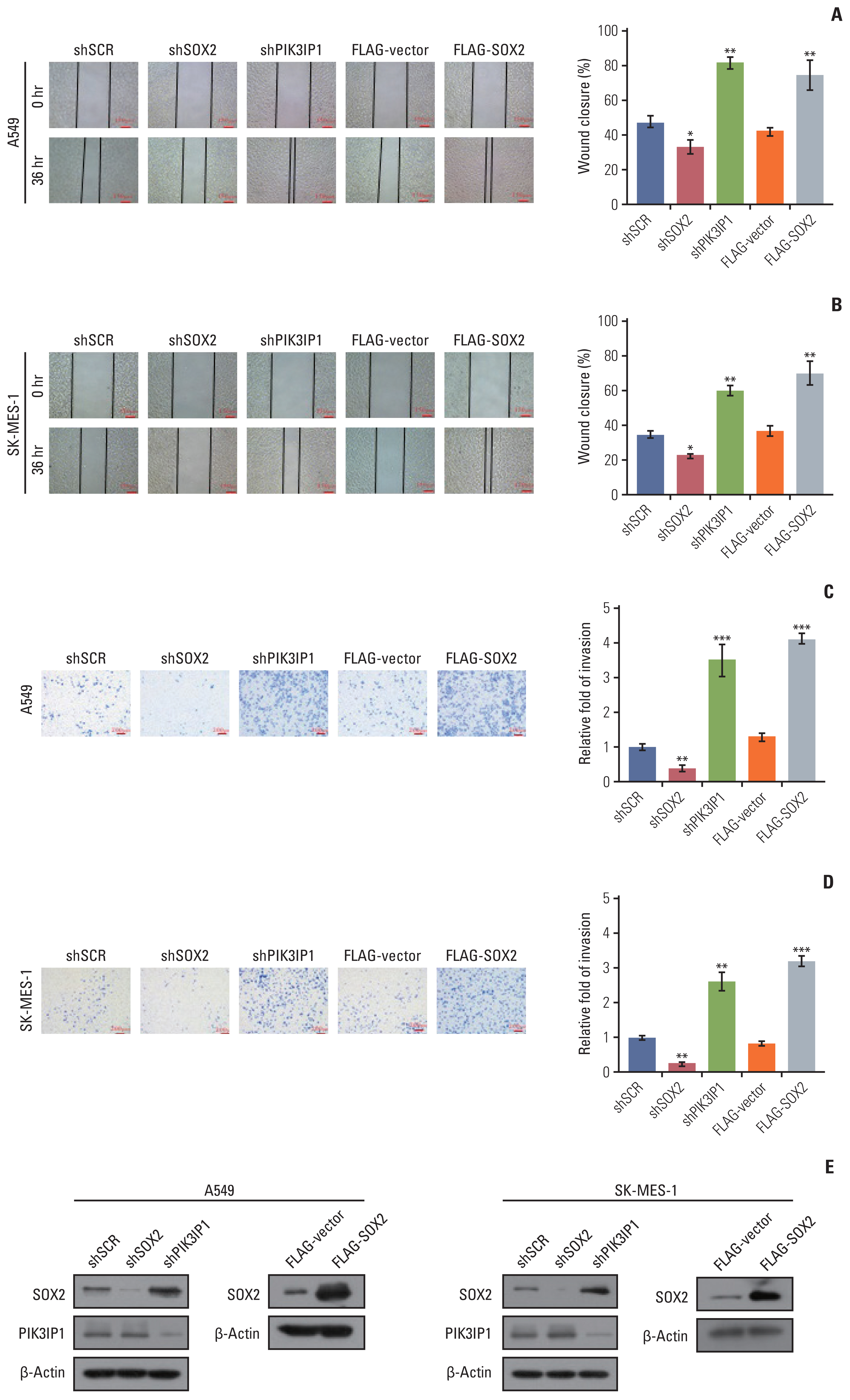
Fig. 7Stable inhibition of mixed-lineage leukemia protein 4 (MLL4) promotes sphere formation of non–small cell lung cancer cells. A549 (A) and SK-MES-1 cells (B) transfected with CRISPR activation (CRISPRa)-Scramble or CRISPRa-MLL4 gRNA or the indicated specific shRNAs or/and expression constructs were plated in the ultra-low attachment 24-well plate at a density of 1,000 cells/well and cultured in tumor sphere medium. Tumor spheres were photographed and counted. **p < 0.01 (two-tailed t test). (C) The activation and knockdown efficiencies of MLL4 were confirmed by quantitative real-time polymerase chain reaction. The overexpression of PIK3IP1 were confirmed by western blot analysis. 
Fig. 8The effect of PIK3IP1 and SOX2 on non–small cell lung cancer cells sphere formation ability. (A, B) A549 and SK-MES-1 cells transfected with the indicated specific shRNAs or/and expression constructs were performed Sphere Formation Assays. Tumor spheres were photographed and counted. (C) Western blot analysis was used to determine the protein expression in these cells using antibodies against the indicated proteins. Each bar represents the mean±standard deviation for triplicate measurements. **p < 0.01 (two-tailed t test). 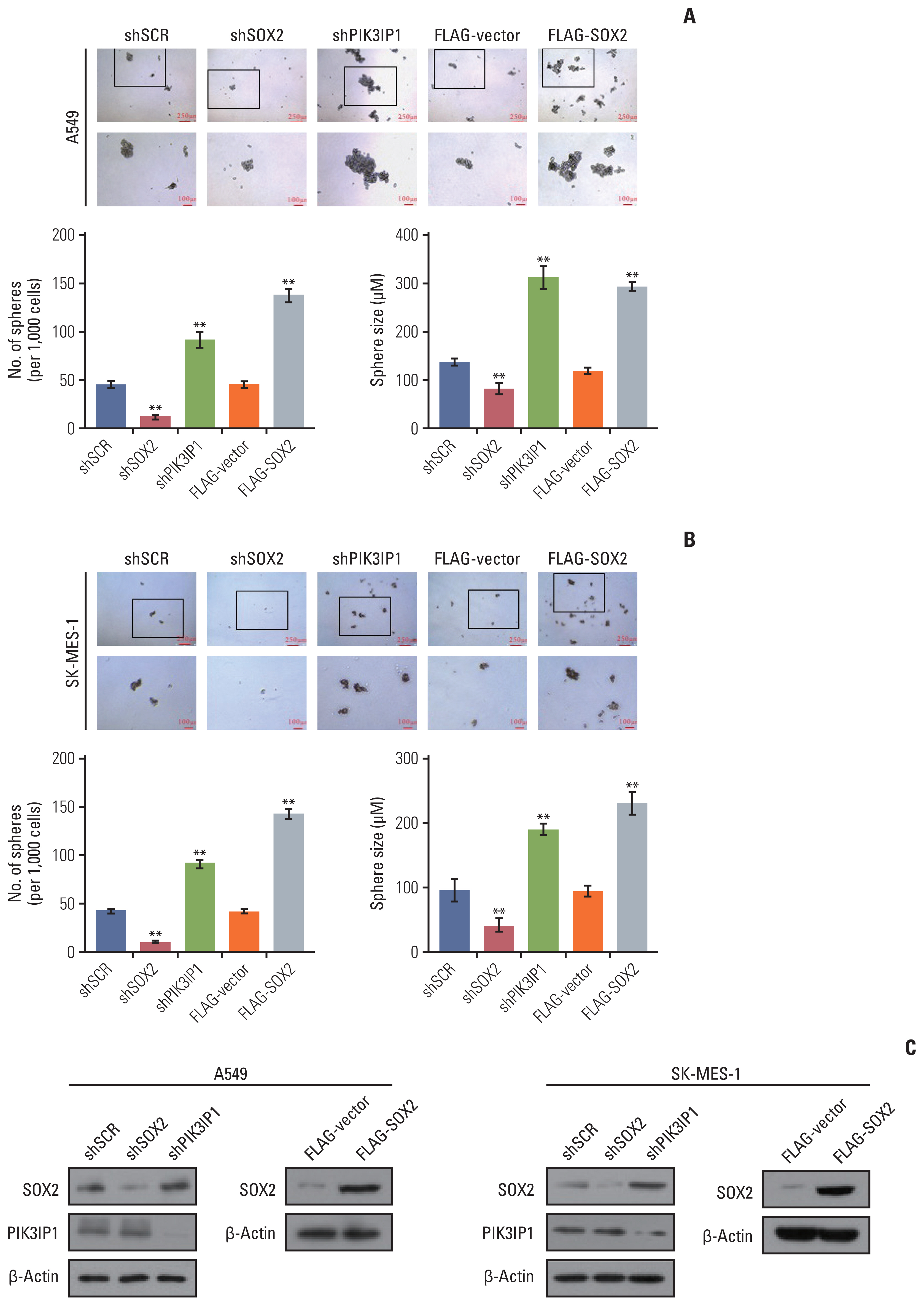
Fig. 9Stable inhibition of mixed-lineage leukemia protein 4 (MLL4) promotes tumorigenesis in vivo. (A) A549 cells stably expressing MLL4 shRNA or shSCR were transplanted into athymic mice. Representative images of tumor-bearing mice and their tumors, the average tumor mass, and weight of each group are shown. **p < 0.01 (two-tailed unpaired t test). (B) A549 cells that had been engineered to stably express firefly luciferase (A549-Luc) were infected with lentiviruses carrying shRNA against MLL4 and then injected intravenously through the tail vein of 6-week-old male NOD SCID mice (n=5). Lung metastasis was quantified using bioluminescence imaging after 4 weeks. Representative in vivo bioluminescent images are shown. Lung cancer specimens were examined by in vitro bioluminescent measurement and the sections from control or shMLL4-treated mice were stained with H&E. (C) Kaplan-Meier survival analysis for the relationship between survival time and the MLL4, PIK3IP1, and SOX2 signature in lung cancer using an online tool (http://kmplot.com/analysis). HR, hazard ratio. (D) Analysis of the correlations between MLL4 and PIK3IP1 and SOX2 in the public datasets GSE108492, GSE115457, GSE8894, and GSE3141. 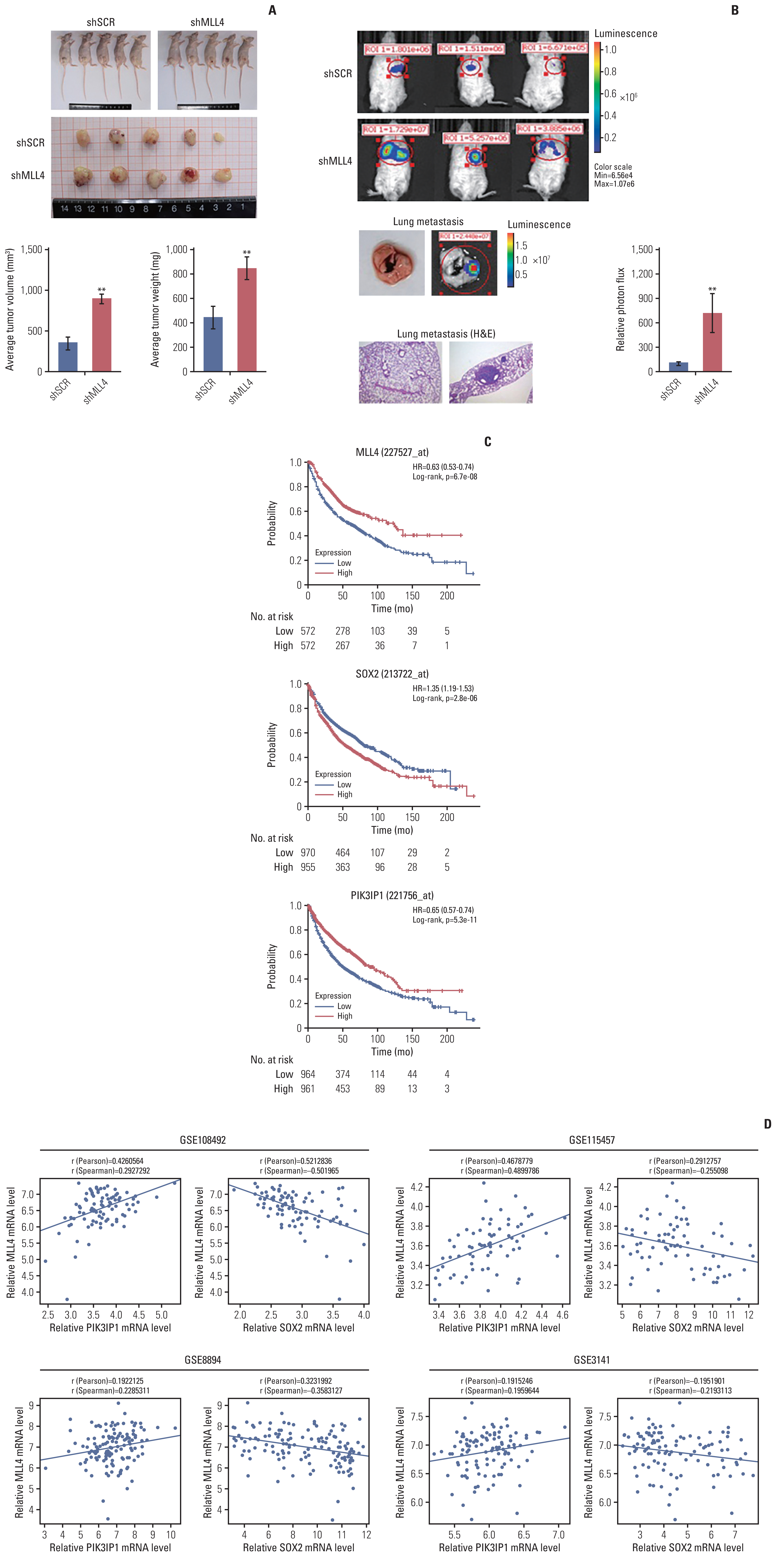
References1. Maiuthed A, Chantarawong W, Chanvorachote P. Lung cancer stem cells and cancer stem cell-targeting natural compounds. Anticancer Res. 2018;38:3797–809.
2. Yang S, Zhang Z, Wang Q. Emerging therapies for small cell lung cancer. J Hematol Oncol. 2019;12:47.
3. Dong S, Li W, Wang L, Hu J, Song Y, Zhang B, et al. Histone-related genes are hypermethylated in lung cancer and hypermethylated HIST1H4F could serve as a pan-cancer biomarker. Cancer Res. 2019;79:6101–12.
4. Fagan RJ, Dingwall AK. COMPASS ascending: emerging clues regarding the roles of MLL3/KMT2C and MLL2/KMT-2D proteins in cancer. Cancer Lett. 2019;458:56–65.
5. Gonda TJ, Ramsay RG. Directly targeting transcriptional dysregulation in cancer. Nat Rev Cancer. 2015;15:686–94.
6. Ford DJ, Dingwall AK. The cancer COMPASS: navigating the functions of MLL complexes in cancer. Cancer Genet. 2015;208:178–91.
7. Ardeshir-Larijani F, Bhateja P, Lipka MB, Sharma N, Fu P, Dowlati A. KMT2D mutation is associated with poor prognosis in non-small-cell lung cancer. Clin Lung Cancer. 2018;19:e489–501.
8. Lin-Shiao E, Lan Y, Coradin M, Anderson A, Donahue G, Simpson CL, et al. KMT2D regulates p63 target enhancers to coordinate epithelial homeostasis. Genes Dev. 2018;32:181–93.
10. Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30.
11. Dawkins JB, Wang J, Maniati E, Heward JA, Koniali L, Kocher HM, et al. Reduced expression of histone methyltransferases KMT2C and KMT2D correlates with improved outcome in pancreatic ductal adenocarcinoma. Cancer Res. 2016;76:4861–71.
12. Lv S, Ji L, Chen B, Liu S, Lei C, Liu X, et al. Histone methyltransferase KMT2D sustains prostate carcinogenesis and metastasis via epigenetically activating LIFR and KLF4. Oncogene. 2018;37:1354–68.
13. Hillman RT, Celestino J, Terranova C, Beird HC, Gumbs C, Little L, et al. KMT2D/MLL2 inactivation is associated with recurrence in adult-type granulosa cell tumors of the ovary. Nat Commun. 2018;9:2496.
14. Sun P, Wu T, Sun X, Cui Z, Zhang H, Xia Q, et al. KMT2D inhibits the growth and metastasis of bladder cancer cells by maintaining the tumor suppressor genes. Biomed Pharmacother. 2019;115:108924.
15. Ortega-Molina A, Boss IW, Canela A, Pan H, Jiang Y, Zhao C, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21:1199–208.
16. Xiong W, Deng Z, Tang Y, Deng Z, Li M. Downregulation of KMT2D suppresses proliferation and induces apoptosis of gastric cancer. Biochem Biophys Res Commun. 2018;504:129–36.
17. Alam H, Tang M, Maitituoheti M, Dhar SS, Kumar M, Han CY, et al. KMT2D deficiency impairs super-enhancers to confer a glycolytic vulnerability in lung cancer. Cancer Cell. 2020;37:599–617.
18. Zeng Y, Qiu R, Yang Y, Gao T, Zheng Y, Huang W, et al. Regulation of EZH2 by SMYD2-mediated lysine methylation is implicated in tumorigenesis. Cell Rep. 2019;29:1482–98.
19. Yang Y, Qiu R, Zhao S, Shen L, Tang B, Weng Q, et al. SMYD3 associates with the NuRD (MTA1/2) complex to regulate transcription and promote proliferation and invasiveness in hepatocellular carcinoma cells. BMC Biol. 2022;20:294.
20. Sikorska M, Sandhu JK, Deb-Rinker P, Jezierski A, Leblanc J, Charlebois C, et al. Epigenetic modifications of SOX2 enhancers, SRR1 and SRR2, correlate with in vitro neural differentiation. J Neurosci Res. 2008;86:1680–93.
21. Ediriweera MK, Tennekoon KH, Samarakoon SR. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: biological and therapeutic significance. Semin Cancer Biol. 2019;59:147–60.
22. Tewari D, Patni P, Bishayee A, Sah AN, Bishayee A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: a novel therapeutic strategy. Semin Cancer Biol. 2022;80:1–17.
23. Yang Q, Jiang W, Hou P. Emerging role of PI3K/AKT in tumor-related epigenetic regulation. Semin Cancer Biol. 2019;59:112–24.
24. Wang Z, Kang L, Zhang H, Huang Y, Fang L, Li M, et al. AKT drives SOX2 overexpression and cancer cell stemness in esophageal cancer by protecting SOX2 from UBR5-mediated degradation. Oncogene. 2019;38:5250–64.
25. Mamun MA, Mannoor K, Cao J, Qadri F, Song X. SOX2 in cancer stemness: tumor malignancy and therapeutic potentials. J Mol Cell Biol. 2020;12:85–98.
26. Karachaliou N, Rosell R, Viteri S. The role of SOX2 in small cell lung cancer, lung adenocarcinoma and squamous cell carcinoma of the lung. Transl Lung Cancer Res. 2013;2:172–9.
27. Singh S, Trevino J, Bora-Singhal N, Coppola D, Haura E, Altiok S, et al. EGFR/Src/Akt signaling modulates Sox2 expression and self-renewal of stem-like side-population cells in non-small cell lung cancer. Mol Cancer. 2012;11:73.
28. Schaefer T, Lengerke C. SOX2 protein biochemistry in stem-ness, reprogramming, and cancer: the PI3K/AKT/SOX2 axis and beyond. Oncogene. 2020;39:278–92.
29. Schaefer T, Ramadoss A, Leu S, Tintignac L, Tostado C, Bink A, et al. Regulation of glioma cell invasion by 3q26 gene products PIK3CA, SOX2 and OPA1. Brain Pathol. 2019;29:336–50.
30. Tang J, Zhong G, Wu J, Chen H, Jia Y. SOX2 recruits KLF4 to regulate nasopharyngeal carcinoma proliferation via PI3K/AKT signaling. Oncogenesis. 2018;7:61.
|
|
|||||||||||||||||||||||||||||||||||||||||||||