AbstractPurposeTransition to next generation sequencing (NGS) for BRCA1/BRCA2 analysis in clinical laboratories is ongoing but different platforms and/or data analysis pipelines give different results resulting in difficulties in implementation. We have evaluated the Ion Personal Genome Machine (PGM) Platforms (Ion PGM, Ion PGM Dx, Thermo Fisher Scientific) for the analysis of BRCA1/2.
Materials and MethodsThe results of Ion PGM with OTG-snpcaller, a pipeline based on Torrent mapping alignment program and Genome Analysis Toolkit, from 75 clinical samples and 14 reference DNA samples were compared with Sanger sequencing for BRCA1/BRCA2. Ten clinical samples and 14 reference DNA samples were additionally sequenced by Ion PGM Dx with Torrent Suite.
ResultsFifty types of variants including 18 pathogenic or variants of unknown significance were identified from 75 clinical samples and known variants of the reference samples were confirmed by Sanger sequencing and/or NGS. One false-negative results were present for Ion PGM/OTG-snpcaller for an indel variant misidentified as a single nucleotide variant. However, eight discordant results were present for Ion PGM Dx/Torrent Suite with both false-positive and -negative results. A 40-bp deletion, a 4-bp deletion and a 1-bp deletion variant was not called and a false-positive deletion was identified. Four other variants were misidentified as another variant.
IntroductionAbout 10% of the breast cancers are known to be hereditary and the most commonly associated gene with hereditary breast and ovarian cancer syndromes (HBOC) are BRCA1 and BRCA2 [1,2]. The detection of genetic variants in HBOC patients and the carriers have enormous impact on the management and decision for further therapeutic options [3]. Thus, performing BRCA1 and BRCA2 analysis is recommended in patients suspected of HBOC [4]. According to the National Comprehensive Cancer Network guidelines, genetic risk assessment and genetic testing is required in patients with pedigrees suggestive of or known genetic predisposition [4]. Sanger sequencing has been the gold standard for BRCA1/BRCA2 variant analysis for HBOC [5,6]. However, due to the relatively large size of the BRCA1 and BRCA2 genes and the lack of mutational hotspots, performing Sanger sequencing in multiple samples is often time consuming and expensive [7]; thus, the implementation of next generation sequencing (NGS) for BRCA1/BRCA2 analysis is being considered in diagnostic settings [8-10]. Many studies have evaluated the use of NGS for the detection of BRCA1 or BRCA2 variants in HBOC [11-16] and some recent studies have evaluated the analysis of BRCA1 or BRCA2 with NGS in the clinical laboratory setting [13,15-17]. In this study, we have assessed the results of NGS assays for BRCA1 and BRCA2 variants performed by an amplicon based method with Ion Torrent Personal Genome Machine (PGM) (Thermo Fisher Scientific, Waltham, MA) and Ion PGM Dx (Thermo Fisher Scientific), in consideration of implementing the platforms in a diagnostic laboratory. Since different platforms and data analysis pipelines give different results, platforms available for diagnostic use were considered. Ion PGM Dx is a class II medical device approved by the U.S. Food and Drug Administration used in conjunction with the instrument-specific reagents and data analysis software, Torrent Suite. Ion Torrent systems measures the pH to detect polymerization events and is known to be prone to homopolymer errors [18,19], thus OTG-snpcaller, an optimized pipeline based on Torrent Mapping Alignment Program (TMAP) and Genome Analysis Tool kit (GATK) [20] for single nucleotide polymorphism calling from Ion Torrent data [21] were used as the analysis pipeline with the Ion PGM data. The results of BRCA1 and BRCA2 analysis with Ion PGM/OTG-snpcaller and Ion PGM Dx/Torrent Suite were compared to those from Sanger sequencing. Since there are no studies with the Ion PGM Dx platform, we have evaluated the results of Ion PGM and Ion PGM Dx for BRCA1/BRCA2 analysis with clinical and reference samples.
Materials and Methods1. SamplesClinical samples were collected from 75 breast cancer patients suspected of hereditary breast cancer for detection of BRCA1 or BRCA2 genetic variants as a routine clinical practice. Seventy-five consecutive samples were included in this study. In addition, 14 reference DNA samples from the National Institute of General Medical Sciences Human Genetic Cell Repository (https://catalog.coriell.org/) with known BRCA1 or BRCA2 pathogenic variants or variants of unknown significance (VUS) were included in this study. The sample numbers used are NA13705, NA13715, NA14090, NA14094, NA14634, NA14636, NA14637, NA14638, NA14684, NA141-70, NA14622, NA14623, NA14624, and NA14639. This study was approved by the Institutional Review Board of the institution (IRB-B-1508-310-302).
2. MethodsThe genomic DNA was extracted by QIAamp Blood DNA mini kit (Qiagen, Valencia, CA) for the clinical samples and the reference DNA samples were obtained. For 75 clinical samples and 14 reference samples, the comparison of results of Sanger sequencing and NGS with Ion PGM was made and among the clinical and reference samples, 24 samples (10 clinical samples and 14 reference samples), were performed additionally with Ion PGM Dx for comparison.
3. Next generation sequencing1) Library preparationThe target regions of BRCA1 and BRCA2 were amplified by the Ion Ampliseq BRCA1 and BRCA2 Panel (Thermo Fisher Scientific). The panel includes three primer pools (167 amplicons) which cover the entire coding region and 10 to 20 bp of the intronic flanking sequences of coding exons. For amplification, 4 μL of 5× Ion Ampliseq HiFi Master Mix (Thermo Fisher Scientific), 10 μL of 2× Ion Ampliseq primer pool, 20 ng of genomic DNA per reaction, and 4 μL of nuclease-free water were mixed. The temperature profile applied with the final 20 μL of the polymerase chain reaction (PCR) mixture were 99ºC for 2 minutes, 99ºC for 15 seconds, and 60ºC for 4 minutes 19 cycles, with a final hold at 10ºC. The primer sequences were partially digested and adapters and barcodes were ligated to the amplicons according to the Ion AmpliSeq Library 2.0 Kit manual. A unique adapter was applied for each library with the Ion Xpress Barcode Adapters 1 to 16 Kit (Thermo Fisher Scientific). Quantification of amplified library was performed with the Qubit ver. 2.0 fluorometer (Life Technologies, Carlsbad, CA) using the Qubit dsDNA HS Assay Kit, diluted to approximately 100 pmol/L. Ion OneTouch 2 System and the Ion OneTouch ES Instrument (Thermo Fisher Scientific) was used according to the user guide with the 200-bp chemistry kits. All barcoded samples were sequenced on the Ion PGM (Thermo Fisher Scientific) with 316 chips using 12 samples on a single chip per sequencing run.
For Ion PGM Dx, Ion PGM Dx Library Kit, Ion OneTouch Dx Template Kit, and the Ion PGM Dx Sequencing Kit was used. All barcoded samples were sequenced on the Ion PGM Dx with the Ion 318 Dx Chip using 12 samples on a single chip per sequencing run. Sequencing data were analyzed with the Ion Torrent Suite software TS 4.0.0 and contextually with Ion Reporter.
2) Data analysis for NGSFor Ion PGM, we have adopted an optimized pipeline based on TMAP and GATK [20], the OTG-snpcaller for single nucleotide variant based on a previous study [21]. Briefly, the raw data from Torrent Suite 4.6 was mapped with TMAP 3.6 (https://github.com/iontorrent/TAMP) and the duplicates were removed with Remove Duplicates according to the Alignment Score Tag. To reduce the false negative results of single nucleotide variant in a gap site, Alignment Optimize Structure filtering method was incorporated. Then the variant calling was performed with GATK tool. Local mutational hotspot files were included for annotation of the variants identified.
For Ion PGM Dx, the sequence data were processed using Ion Torrent Suite software 4.0 processed on the Torrent Server (Thermo Fisher Scientific).
Sequence alignments for variants with discordant results were manually inspected with the Integrative Genomics Viewer (IGV) 2.3 [22].
4. Sanger sequencingAll clinical samples were sequenced for the entire coding regions by Sanger sequencing. B-Pure EasySeq PCR plates for BRCA1 and BRCA2 (Nimagen, Nijmegen, Netherlands) were used for amplification and the PCR products were purified and sequenced with BigDye Terminator Cycle Sequencing Kit (Applied Biosystem, Foster City, CA) and products were run on 3130xl Genetic Analyzer (Applied Biosystems). The data was analyzed by Mutation Surveyor 4.0 (SoftGenetics, State College, PA) with the reference sequence NM_007294.2 for BRCA1 and NM_000059.3 for BRCA2. The variants were described as recommended by the Human Genome Variation Society (HGVS) nomenclature (http://varnomen.hgvs.org).
The characterization of the variants was made primarily by Breast Cancer Information Core (BIC) from National Human Genome Institute (https://research.nhgri.nih.gov/bic/), Short Genetic Variations database (dbSNP, https://www.ncbi.nlm.nih.gov/projects/SNP/index.html), and Human Gene Mutation Database (HGMD) professional 2016.2 (Qiagen, Boston, MA) as of date September 10, 2016. BIC classifies variants into five classes: class 1 for not pathogenic/low clinical significance, class 2 for likely not pathogenic/little clinical significance, class 3 for uncertain variants, class 4 for likely pathogenic variants, and class 5 for pathogenic variants. HGMD classifies variants into disease causing mutation, likely disease causing mutation, disease associated polymorphism, disease associated polymorphism with additional supporting functional evidence, frameshift or truncating variant with no disease association reported yet, polymorphism affecting the structure, function or expression of a gene but with no disease association reported yet.
5. Multiple ligation probe amplificationTo analyze for large gene rearrangement or deletion/duplication, multiple ligation probe amplification (MLPA) was performed for all clinical samples. MLPA kits (P002 for BRCA1 and P045 for BRCA2) (MRC-Holland, Amsterdam, Netherlands) were used with Veriti 96-well Thermal Cycler (Applied Biosystems) and the data was analyzed with GeneMarker 2.0 (SoftGenetics).
Results1. Sequencing (NGS) statisticsFor BRCA1 and BRCA2 analysis, on average, 233,185 reads per patients were obtained with a mean amplicon length of 139 base pair (bp) with Ion PGM in 89 samples. The mean sequencing depth was 1,377× (775× to 2,237×) and 96.40% of the reads were on the targeted region of BRCA1 and BRCA2. The uniformity of coverage was 97.22%. Twenty-four samples were performed concurrently with Ion PGM Dx and the mean read length was 149 bp, the mean mapped reads were 280,200 bp and on target rate was 97.80% with a mean depth of 1,796× (1,062× to 4,332×) and the mean uniformity of 96.23%.
2. Detection of variantsPathogenic variants and VUS identified in 75 clinical samples and reference DNA samples are shown in Tables 1 and 2, respectively. MLPA results showed no deletions or duplications for BRCA1 and BRCA2 in clinical samples. In clinical samples, six pathogenic or VUS were found in BRCA1 including two nonsense variants, one frameshift variant, and three missense variants. Twelve pathogenic or VUS were identified in BRCA2 including seven frameshift and five missense variants. All pathogenic or VUS identified in the clinical samples were previously reported variants. A list of all variants (n=50) identified in clinical samples are shown in the S1 Table. In total, 19 variants were identified in BRCA1 including 18 exonic and one intronic variants. Six synonymous, nine missense variants, two nonsense variants, and one frameshift variants were identified in the exons. In BRCA2, 31 variants were identified with 30 exonic variants and one intronic variant. There were seven frameshift variants, 13 missense and 10 synonymous variants. In total, 14 pathogenic/VUS variants were found in the reference DNA samples in this study, which included nine variants in BRCA1 (one nonsense, one splice site, and seven frameshift variants) and five variants in BRCA2 including one missense and four frameshift variants.
The variants identified by the NGS and Sanger sequencing were compared. The different parameters for variant calling in Ion Torrent variant caller for Ion PGM Dx are shown in S2 Table. In Ion PGM, the “Generic-PGM-Germline Low stringency” parameter was used for variant calling while the default option was used for Ion PGM Dx/Torrent Suite.
The discordant sequencing results including those from clinical and reference DNA samples are shown in Table 3. Ion PGM/OTG-snpcaller and Sanger sequencing showed only one discrepant result in BRCA1 for c.922_924delinsT in which Ion PGM/OTG-snpcaller identified the indel as a single nucleotide variant, c.922A>T (Fig. 1). All the other pathogenic variants or VUS identified were in concordance with Sanger sequencing. However, eight discrepant results were present between Sanger sequencing and Ion PGM Dx/Torrent Suite. One false-positive variant in BRCA1, c.117_118delTG was identified by Ion PGM Dx but not present by visual inspection with the IGV. Three false-negative results were present with Ion PGM Dx/Torrent Suite in which pathogenic variants of BRCA1, c.1175_1214del40, c.4065_4068delTCAA and a pathogenic variant of BRCA2, c.994delA were not called by the Ion PGM Dx/Torrent Suite (Fig. 2). Four other discordant results were present misidentifying a variant as another variant with Ion PGM Dx/Torrent Suite and of these discordant results, two variants were recurrently present in seven samples among 24 samples tested with Ion PGM Dx/Torrent Suite. Single nucleotide variants of BRCA1, c.3113A>G and c.3548A>G, were called as c.3107_3112delTTAAAG and c.3548_3549delAA. The other misidentified variant was c.922_924delinsT which was called as c.922A>T as in Ion PGM/OTG-snpcaller and a 4-bp deletion in BRCA2, c.3741_3744delTGAG, which was c.3744_3747delAGTG by Sanger sequencing.
DiscussionThe implementation of NGS to a clinical laboratory requires validation and thorough evaluation. Many studies have shown comparison results of BRCA1 and BRCA2 with NGS to Sanger sequencing, which has been considered the golden standard for variant analysis. However, different NGS platforms and data analysis pipeline showed variable performance [12,13,17,23], which lead us to the evaluation of Ion PGM and Ion PGM Dx, which were candidates of NGS platforms to our laboratory. Our data showed that Ion PGM/OTG-snpcaller showed comparable results to Sanger sequencing with one false negative results for an insertion/deletion variant. However, Ion PGM Dx with the supplied Torrent Suite software alone was not suitable for BRCA1 and BRCA2 analysis in a diagnostic setting, due to false negative and positive errors.
In total, 50 variants including 12 pathogenic variants/VUS and 38 benign variants were found in 75 clinical samples and the known pathogenic or VUS of the 14 reference Coriell DNA samples were confirmed with Sanger sequencing and NGS. Most of the variants were identified with Sanger sequencing and Ion PGM/OTG-snpcaller except for one discordant result for a pathogenic variant. An indel variant in BRCA1, c.922_924delinsT was identified as a single nucleotide variant (c.922A>T) with Ion PGM/OTG-snpcaller and this variant was also not correctly called by Ion PGM Dx/Torrent Suite.
Several other discrepancies were present with Ion PGM Dx/Torrent Suite, showing recurrent false positive and negative variant assignments. Three recurrent false positive variants in BRCA1, c.3107_3112delTTAAAG and c.3548_3549delAA, which corresponded to c.3113A>G and c.3548A>G by Sanger sequencing were present in the variant call format (VCF), however there were no insertions or deletions manually confirmed by IGV. We have speculated that these false positive assignments occur when multiple possible variants are listed for a chromosome position which includes a hotspot variant inserted prior to analysis. Therefore, to minimize the discrepancies by Ion PGM Dx/Torrent Suite, we have deleted the two recurrent hotspot variants from the local database and those recurrently misidentified as a pathogenic variant were not reported in the VCF files. However, the manual deletions of these hotspot variants would be not advisable in populations where these variants are reported. Moreover, since incorrect hotspot IDs were matched to the variants, we have developed a simple program with python 2.7, “Filter Dx” which selects homozygous or heterozygous variants and eliminates the assigned hotspot variants when there are multiple alternative alleles, where errors were found to be prone. This program condenses the long list of variants with possible germline variants (provided at request).
False-negative results were present by Ion PGM Dx missing a 40-bp deletion in BRCA1 (c.1175_1214del40), a 4-bp deletion in BRCA1(c.4065_4068delTCAA) in reference DNA samples and a 1-bp deletion in BRCA2 (c.994delA) (Fig. 2). The 40-bp deletion have been reported as being difficult in NGS for detection and not detected in certain platforms requiring modification of data analysis pipelines [17,24,25]. Not many parameters were adjustable in the Ion PGM Dx/Torrent Suite platform in processing the data acquired from the Ion PGM Dx, thus requiring further modification of the variant calling pipelines in Torrent Suite as suggested in previous studies for its use in the clinical laboratory setting [17,26].
As in a previous study, we have also found that Ion PGM Dx/Torrent Suite reported a variant with AGTG deletion from chr13:32912234 (c.3742_3745del), whereas a deletion of TGAG from chr13:32912236, c.3744_3747delTGAG was detected in Sanger sequencing [16] and the discrepancy in codon numbers occurs since the HGVS nomenclature requires that the most 3’ position possible of the reference sequence is arbitrarily assigned to have been changed. Thus, confirming the variants with IGV and checking the HGVS nomenclature is essential for reporting variants.
Ion Torrent platforms is known to have disadvantages in homopolymer errors, false indel detections [19,26]; however, many studies have shown that Ion Torrent platforms can be used in clinical settings when sufficient read depth can be obtained and quality control measures are implemented [8,12,13]. In our study, modifying the data analysis algorithm with TMAP and GATK (OTG-snpcaller) with Ion PGM reduced the calling errors and identified the variants not called by Ion PGM Dx/Torrent Suite, showing the importance of optimizing data analysis pipeline for implementing NGS [12,26].
The reference DNA samples allowed confirmation of pathogenic variants with the various methods, helping the evaluation of procedural evaluations as incorporated in many of the validation studies with NGS [24,25,27].
Although approved as class II medical device, the use of Ion PGM Dx with the vendor supplied Torrent Suite analysis software showed both false-negative and -positive results not suitable for use in a clinical laboratory. Further optimization of the data analysis pipeline is necessary for use of Ion PGM platforms to be used in a clinical laboratory.
Electronic Supplementary MaterialSupplementary materials are available at Cancer Research and Treatment website (http://www.e-crt.org).
Fig. 1.Alignment of an indel variant (c.922_924delinsT) in a clinical sample viewed by Integrative Genomics Viewer, called as a single nucleotide variant, c.922C>T, by both Ion PGM/OTG-snpcaller and Ion PGM Dx/Torrent Suite. 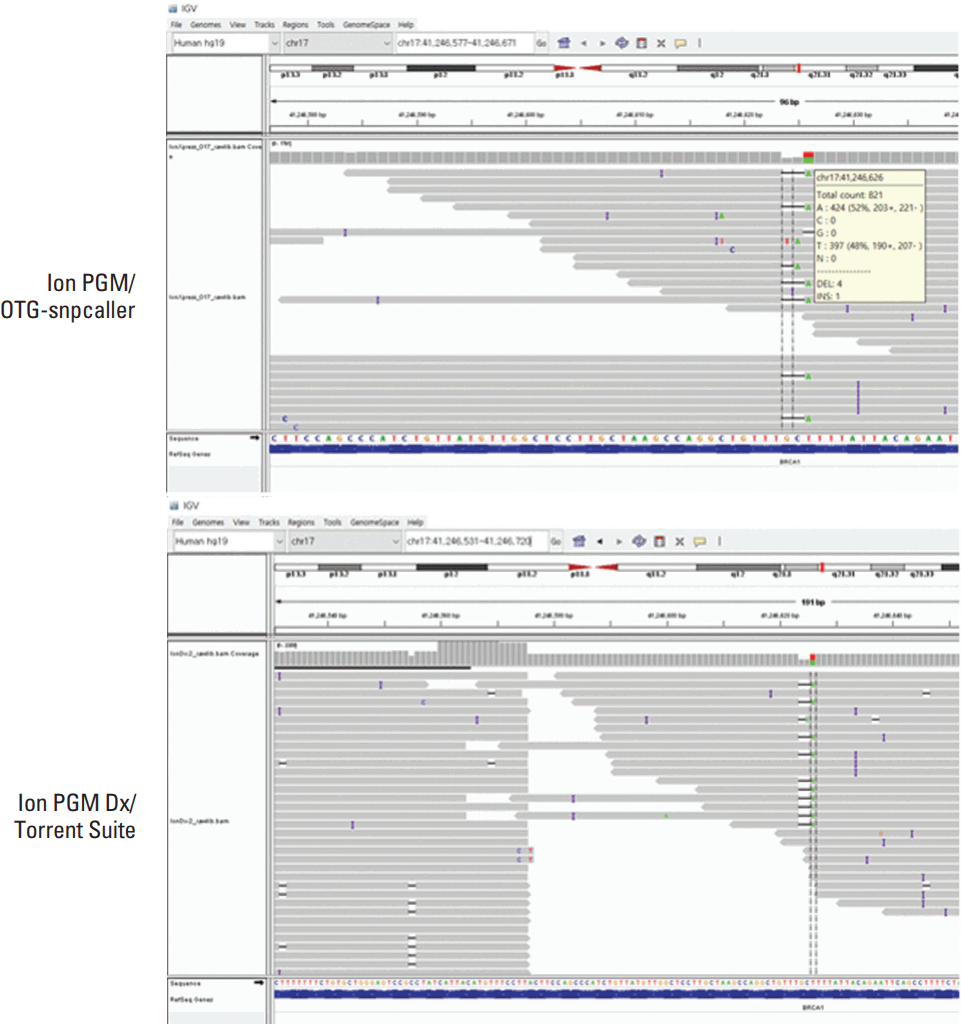
Fig. 2.(A) Alignment of a 40-bp deletion in BRCA1 (c.1175_1214del40) in the reference Coriell DNA sample (NA14094) which was called by Ion PGM/OTG-snpcaller but not by Ion PGM Dx/Torrent Suite. (B) A 4-bp deletion in BRCA1 (c.4065_4068delTCAA) not called by Ion PGM Dx/Torrent Suite present in a reference Coriell DNA sample (NA14634). (C) A 1-bp deletion in BRCA2 (c.994delA) not called by Ion PGM Dx/Torrent Suite present in a clinical sample. 
Table 1.List of pathogenic variants and variants of known significance found in clinical samples Table 2.List of reference Coriell DNA samples and the pathogenic/unknown significance variants in each sample Table 3.Discordant results by NGS and Sanger sequencing References1. Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–89.
2. King MC, Marks JH, Mandell JB; New York Breast Cancer Study Group. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–6.
3. Hartmann LC, Lindor NM. The role of risk-reducing surgery in hereditary breast and ovarian cancer. N Engl J Med. 2016;374:454–68.
4. National Comprehensive Cancer Network. NCCN guidelines, version 2. 2016. Breast and/or ovarian cancer genetic assessment. Fort Washington, PA: National Comprehensive Cancer Network; 2016.
5. Tafe LJ, Datto MB, Palomaki GE, Lacbawan FL; CAP/ACMG Biochemical and Molecular Genetics Resource Committee. Molecular testing for the BRCA1 and BRCA2 Ashkenazi Jewish founder mutations: a report on the College of American Pathologists proficiency testing surveys. Genet Med. 2015;17:58–62.
6. Gerhardus A, Schleberger H, Schlegelberger B, Gadzicki D. Diagnostic accuracy of methods for the detection of BRCA1 and BRCA2 mutations: a systematic review. Eur J Hum Genet. 2007;15:619–27.
7. Chan M, Ji SM, Yeo ZX, Gan L, Yap E, Yap YS, et al. Development of a next-generation sequencing method for BRCA mutation screening: a comparison between a high-throughput and a benchtop platform. J Mol Diagn. 2012;14:602–12.
8. Feliubadalo L, Lopez-Doriga A, Castellsague E, del Valle J, Menendez M, Tornero E, et al. Next-generation sequencing meets genetic diagnostics: development of a comprehensive workflow for the analysis of BRCA1 and BRCA2 genes. Eur J Hum Genet. 2013;21:864–70.
9. Judkins T, Leclair B, Bowles K, Gutin N, Trost J, McCulloch J, et al. Development and analytical validation of a 25-gene next generation sequencing panel that includes the BRCA1 and BRCA2 genes to assess hereditary cancer risk. BMC Cancer. 2015;15:215.
10. De Leeneer K, Hellemans J, De Schrijver J, Baetens M, Poppe B, Van Criekinge W, et al. Massive parallel amplicon sequencing of the breast cancer genes BRCA1 and BRCA2: opportunities, challenges, and limitations. Hum Mutat. 2011;32:335–44.
11. Bosdet IE, Docking TR, Butterfield YS, Mungall AJ, Zeng T, Coope RJ, et al. A clinically validated diagnostic second-generation sequencing assay for detection of hereditary BRCA1 and BRCA2 mutations. J Mol Diagn. 2013;15:796–809.
12. Tarabeux J, Zeitouni B, Moncoutier V, Tenreiro H, Abidallah K, Lair S, et al. Streamlined ion torrent PGM-based diagnostics: BRCA1 and BRCA2 genes as a model. Eur J Hum Genet. 2014;22:535–41.
13. Dacheva D, Dodova R, Popov I, Goranova T, Mitkova A, Mitev V, et al. Validation of an NGS approach for diagnostic BRCA1/BRCA2 mutation testing. Mol Diagn Ther. 2015;19:119–30.
14. Kluska A, Balabas A, Paziewska A, Kulecka M, Nowakowska D, Mikula M, et al. New recurrent BRCA1/2 mutations in Polish patients with familial breast/ovarian cancer detected by next generation sequencing. BMC Med Genomics. 2015;8:19.
15. Trujillano D, Weiss ME, Schneider J, Koster J, Papachristos EB, Saviouk V, et al. Next-generation sequencing of the BRCA1 and BRCA2 genes for the genetic diagnostics of hereditary breast and/or ovarian cancer. J Mol Diagn. 2015;17:162–70.
16. Park J, Jang W, Chae H, Kim Y, Chi HY, Kim M. Comparison of targeted next-generation and Sanger sequencing for the BRCA1 and BRCA2 mutation screening. Ann Lab Med. 2016;36:197–201.
17. Strom CM, Rivera S, Elzinga C, Angeloni T, Rosenthal SH, Goos-Root D, et al. Development and validation of a next-generation sequencing assay for BRCA1 and BRCA2 variants for the clinical laboratory. PLoS One. 2015;10:e0136419
18. Yeo ZX, Chan M, Yap YS, Ang P, Rozen S, Lee AS. Improving indel detection specificity of the Ion Torrent PGM benchtop sequencer. PLoS One. 2012;7:e45798
19. Bragg LM, Stone G, Butler MK, Hugenholtz P, Tyson GW. Shining a light on dark sequencing: characterising errors in Ion Torrent PGM data. PLoS Comput Biol. 2013;9:e1003031
20. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
21. Zhu P, He L, Li Y, Huang W, Xi F, Lin L, et al. OTG-snpcaller: an optimized pipeline based on TMAP and GATK for SNP calling from ion torrent data. PLoS One. 2014;9:e97507
22. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6.
23. Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341.
24. Enyedi MZ, Jaksa G, Pinter L, Sukosd F, Gyuris Z, Hajdu A, et al. Simultaneous detection of BRCA mutations and large genomic rearrangements in germline DNA and FFPE tumor samples. Oncotarget. 2016;7:61845–59.
25. Lincoln SE, Kobayashi Y, Anderson MJ, Yang S, Desmond AJ, Mills MA, et al. A systematic comparison of traditional and multigene panel testing for hereditary breast and ovarian cancer genes in more than 1000 patients. J Mol Diagn. 2015;17:533–44.
|
|
|||||||||||||||||||||||||||||||||||||||||||||