AbstractPurposeThis study was conducted to investigate the role of four polymorphic variants of DNA methyltransferase genes as risk factors for radiation-induced fibrosis in breast cancer patients. We also assessed their ability to improve prediction accuracy when combined with mitochondrial haplogroup H, which we previously found to be independently associated with a lower hazard of radiation-induced fibrosis.
Materials and Methods
DNMT1 rs2228611,DNMT3A rs1550117,DNMT3A rs7581217, and DNMT3B rs2424908 were genotyped by real-time polymerase chain reaction in 286 Italian breast cancer patients who received radiotherapy after breast conserving surgery. Subcutaneous fibrosis was scored according to the Late Effects of Normal Tissue–Subjective Objective Management Analytical (LENT-SOMA) scale. The discriminative accuracy of genetic models was assessed by the area under the receiver operating characteristic curves (AUC).
ResultsKaplan-Meier curves showed significant differences among DNMT1 rs2228611 genotypes in the cumulative incidence of grade ≥ 2 subcutaneous fibrosis (log-rank test p-value= 0.018). Multivariate Cox regression analysis revealed DNMT1 rs2228611 as an independent protective factor for moderate to severe radiation-induced fibrosis (GG vs. AA; hazard ratio, 0.26; 95% confidence interval [CI], 0.10 to 0.71; p=0.009). Adding DNMT1 rs2228611 to haplogroup H increased the discrimination accuracy (AUC) of the model from 0.595 (95% CI, 0.536 to 0.653) to 0.655 (95% CI, 0.597 to 0.710).
IntroductionAdjuvant radiotherapy (RT) after conservative surgery for early stage breast cancer is a standard of care that is well tolerated by the patients in terms of compliance and risk of adverse effects. In most cases, breast cancer patients have a long life expectancy with a risk of late normal tissue complications that may continue to accumulate years after treatment concludes. For this reason, late effects remain important health concerns for long-term survivors [1]. Common late toxicity manifestations in the breast include fibrosis, cutaneous atrophy and skin telangiectasia [2]. In particular, fibrosis is the development of excess fibrous connective tissue due to fibroblasts proliferation because of tissue injury. This leads to breast indurations, and in some cases to unfavorable cosmetic outcome [3]. There is considerable interindividual variability in the development of late toxicity, which depends on RT parameters such as total dose, dose per fraction, irradiated volume, and dose inhomogeneity [4], as well as on patient related factors such as age and life style factors [5]. In the last decade, several studies have suggested that development of late normal tissue complications in the breast after RT is at least in part genetically determined [6]. Currently, clinical radiosensitivity is nowadays regarded as a complex polygenic trait resulting from the combined effects of multiple factors, each with relatively modest effects [7]. Therefore, an approach based on the combination of multiple genetic factors into a genetic risk score (GRS) may be an attractive strategy for prediction of adverse RT effects.
DNA methylation is an epigenetic mechanism established and maintained by a family of DNA methyltransferase (DNMT) enzymes that catalyze the addition of a methyl group to cytosine residues, using S-adenosylmethionine as the methyl group donor [8]. DNA methylation generally occurs on cytosine residues located in CpG dinucleotides within the promoter region, resulting in chromatin compaction and gene silencing [9]. Three active DNMTs have been described in mammals: DNMT1 is required for maintenance of methylation through generations, while DNMT3A and DNMT3B are mainly involved in establishment of de novo DNA methylation patterns [10]. In vitro and in vivo experimental evidence suggests that enzymes of the methylation machinery play a role in fibrogenesis and radiation response. For instance, upregulation of DNMT1 has been detected in the fibrotic tissue of the skin, kidneys, lungs, and liver [11], whereas activation of myofibroblasts or hepatic stellate cells can be reversed via inhibition of DNMT1 by DNA-demethylating drugs or by specific siRNA shutdown [12]. In addition, reduction of global methylation levels has been reported after irradiation, probably due to decreased expression of DNMT1, DNMT3A, and DNMT3B [13]. Despite evidence of involvement of DNA methylating enzymes in fibrogenesis and radiation response, no information is currently available regarding whether common genetic variants of DNMTs genes contribute to the development of radiation-induced fibrosis in cancer patients. However, recent in vitro studies suggest that mitochondria are the primary loci of RT effects [14], and that mitochondrial DNA haplogroups differently affect mRNA expression of DNMT1, DNMT3A, and DNMT3B, [15] as well as global DNA methylation levels [16].
In the present study, we assessed the role of four single nucleotide polymorphisms (SNPs) of DNMT genes (DNMT1 rs2228611, DNMT3A rs1550117, DNMT3A rs7581217, and DNMT3B rs2424908) as risk factors for subcutaneous fibrosis in a cohort of Italian breast cancer patients who received RT after breast conserving surgery. In addition to DNMT SNPs, we evaluated the predictive role of XRCC1 rs2682585, which was previously reported to be associated with the Standardized Total Average Toxicity (STAT) score, an index of overall toxicity combining skin toxicities and fibrosis of the breast [17]. We also assessed the ability of the aforementioned SNPs to improve prediction accuracy when combined with mitochondrial haplogroup H, which we recently found to be independently associated with a lower hazard of radiationinduced fibrosis in breast cancer patients [18].
Materials and Methods1. Study subject and data collectionThis study included 286 Caucasian patients affected by histologically confirmed breast cancer who underwent conservative surgery and adjuvant RT from 1989 to 2010 at our Department of Radiotherapy. Study details were described in full in our prior publication [18]. Briefly, RT consisted of two opposite tangential wedged beams, followed by a boost on the tumor bed. Radiation therapy was planned on computed tomography slices in all cases. Patients underwent whole breast RT with conventional fractionation to a total dose of 50 Gy followed by boost dose on the tumor bed in cases of invasive tumors. At the time of patient recruitment, a peripheral blood sample was taken and stored at 4°C until analysis. During annual follow-up visits (last update on January 2015), radiation oncologists evaluated the appearance of subcutaneous and cutaneous late toxicities, with particular attention to the onset of fibrosis. Toxicity was scored according to the Late effects of Normal Tissue-Subjective Objective Management Analytical (LENT-SOMA) [19] scale. Patients with moderate to severe fibrosis (≥ grade 2) were referred to as the "radiosensitive group" and compared to patients with no or minimal fibrotic reactions (grade 0-1, control group). This study was approved by the local Ethics Committees of our University Hospital and met the requirements of the Declaration of Helsinki. Informed consent was obtained from all patients before participation in the study.
2. GenotypingDetermination of SNPs was conducted on genomic DNA by real-time polymerase chain reaction (PCR) using the following TaqMan Pre-Designed SNP Genotyping assays (Applied Biosystems, Milan, Italy): C_27838930_10 (DNMT1 rs2228611); C_8722920_10 (DNMT3A rs1550117); C_7863728_10 (DNMT3A rs7581217); C_16013055_10 (DNMT3B rs2424908); and C_16269889_10 (XRCC1 rs2682585). Real-time PCR amplification and detection was performed in 96-well PCR plates using a CFX Connect Real-Time PCR Detection System (Bio-Rad, Milan, Italy).
3. Statistical analysisEach polymorphism was tested for deviation from the Hardy-Weinberg equilibrium (HWE) by use of Pearson’s chisquared test as implemented in Finetti’s program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). For the selected polymorphisms, we considered the co-dominant, dominant, and recessive modes of inheritance. The time to event end-point (grade ≥ 2 fibrosis) was calculated from the first session of RT, and patients not experiencing the end-point were censored at the last follow-up performed. The cumulative incidence of grade ≥ 2 fibrosis was calculated by the Kaplan-Meier method and comparisons between genotype groups were performed using the log-rank test. Univariate Cox regression analyses were performed to calculate the hazard ratio (HR) and the 95% confidence interval (CI) to evaluate the influence of genotypes on grade ≥ 2 fibrosis risk. Multivariate adjustments were made for the mitochondrial haplogroup (H vs. non-H) and clinical variables having a cut-off p-value of < 0.20 in univariate Cox analysis (body mass index, breast diameter, adjuvant treatment, dose per fraction, radiation quality, acute skin toxicity, and postsurgical complications) [18]. To avoid the risk of over-fitting biases [20], only significant genetic factors were considered for construction of a GRS. To accomplish this, the Cox proportional hazard regression coefficient of each genetic factor was converted into an integer risk score by rounding the quotient and dividing the regression coefficient by a single constant. The final risk score represented the sum of integer coefficients. Discrimination capabilities of genetic models were assessed using the area under the receiver operating characteristic curve (ROC). The area under the ROC curve and comparisons between ROC curves were calculated using the method described by DeLong et al. [21]. All statistical analyses were performed using MedCalc ver. 13.3.3 (MedCalc Software, Mariakerke, Belgium) software. Because of the exploratory nature of this study, we reported nominal statistical associations (p < 0.05). Adjusted p-values based on the Bonferroni correction were also considered to avoid chance findings due to multiple testing of five SNPs, and the significance was lowered to p < 0.01.
Results1. Single locus analysisOverall, 51 of the 286 participants (17.8%) experienced moderate to severe fibrosis (LENT-SOMA ≥ grade 2), while 235 patients (82.2%) had no or minimal fibrosis (LENTSOMA grade 0-1). Detailed demographic and clinical data in the whole cohort of breast cancer patients and following stratification according to their radiosensitive status have been reported elsewhere [18]. The distributions of genotype frequencies for the polymorphisms analyzed were in HWE (all p > 0.05). For the entire set of breast cancer patients, frequencies of minor variant alleles were 0.48 (DNMT1 rs2228611G), 0.09 (DNMT3A rs1550117A), 0.35 (DNMT3A rs7581217T), 0.14 (DNMT3B rs2424908T), and 0.22 (XRCC1 rs2682585). Kaplan-Meier curves for DNMT1 rs2228611 showed differences among genotypes in the cumulative incidence of grade ≥ 2 subcutaneous fibrosis (log-rank test p-value=0.018) (Fig. 1). Univariate Cox regression analysis revealed that DNMT1 rs2228611 was associated with a lower risk of grade ≥ 2 fibrosis under either the codominant (GG vs. AA: HR, 0.26; 95% CI, 0.10 to 0.70; p=0.007) or the recessive contrast (GG vs. AA+AG: HR, 0.31; 95% CI, 0.12 to 0.78; p=0.013) (Table 1). Multivariate Cox regression analysis adjusted for mitochondrial haplogroup (H vs. non-H) and clinical confounding factors revealed DNMT1 rs2228611 as an independent protective factor for moderate to severe radiation-induced fibrosis (GG vs. AA: HR, 0.26; 95% CI, 0.10 to 0.71; p=0.009; GG vs. AA+AG: HR, 0.29; 95% CI, 0.12 to 0.75; p=0.011) (Table 1). Conversely, none of the other DNMT SNPs (Table 1), nor XRCC1 rs2682585 (Table 2) were associated with radiation-induced fibrosis of the breast upon both univariate or multivariate Cox regression analysis. It is worth noting that mitochondrial haplogroup H emerged as a significant protective factor in these multivariate models when DNMT1 rs2228611 was considered under the codominant (HR, 0.48; 95% CI, 0.26 to 0.88; p=0.018) or the recessive mode of inheritance (HR, 0.48; 95% CI, 0.26 to 0.89; p=0.019).
2. GRS analysisTo understand the joint effects of genetic markers on individual risk of grade ≥ 2 fibrosis, we built a GRS for grade ≥ 2 fibrosis based on different combinations of DNMT1 rs2228611 and/or mitochondrial haplogroups according to five Cox regression models, as shown in Table 3. Model 1 included mitochondrial haplogroups only (H vs. non-H), model 2 and model 3 comprised the codominant or the recessive contrast of rs2228611, respectively, model 4 combined mitochondrial haplogroup H with the codominant contrast of DNMT1 rs2228611 and model 5 incorporated mitochondrial haplogroup H with the recessive contrast of DNMT1 rs2228611. We then performed ROC curve analyses to determine the performance of the five models at discriminating patients with or without grade ≥ 2 fibrosis (Fig. 2). The lowest discrimination accuracy, quantified by area under the receiver operating characteristic curves (AUC), was obtained for models 2 and 3, which were based on the two contrasts of rs2228611 (both AUC, 0.587; 95% CI, 0.28 to 0.645), and the highest was observed for model 5, which included the combination of mitochondrial haplogroup H with the recessive contrast of rs2228611 (AUC, 0.655; 95% CI, 0.597 to 0.710). Thus, based on the results shown in Fig. 2, adding the recessive contrast of DNMT1 rs2228611 to haplogroup H determined an increase of the discrimination accuracy from 0.595 (95% CI, 0.536 to 0.653) to 0.655 (95% CI, 0.597 to 0.710). When applying the score developed in model 5, the proportion of patients with grade ≥ 2 radiation-induced fibrosis for each score group showed an increasing trend from lower to higher sum risk scores: 3.4% (score 0), 10.0% (score 1), 14.0% (score 2), and 27.3% (score 2). The Kaplan-Meier analysis of model 5 revealed that patients with an increasing sum risk score showed a trend towards higher frequencies of grade ≥ 2 fibrosis (p trend=0.0005) (Fig. 3).
DiscussionIn the present study, we tested whether the occurrence of radiation-induced fibrosis in breast cancer patients could be at least partially explained by genetic variability in DNMT genes, which encode for epigenetic enzymes known to be involved in fibrogenesis and radiation response [11,12]. To accomplish this, we investigated the association between common polymorphisms in DNMT1, DNMT3A, and DNMT3B genes and the risk of subcutaneous fibrosis in a cohort of breast cancer survivors who received RT after breast-conserving surgery. Multivariate Cox analysis revealed DNMT1 rs2228611GG genotype as an independent predictor of lower risk for radiation-induced skin fibrosis. To the best of our knowledge, this is the first study to suggest that genetic variability at the DNMT1 locus contributes to the development of late normal tissue complications after radiation therapy, which further supports the importance of DNMT1 on fibrogenesis and wound healing after radiation exposure [12]. It is worth noting that XRCC1 rs2682585, previously reported to be associated with an overall toxicity score combining skin toxicities and fibrosis of the breast [17], was not found to be correlated with fibrosis alone in the present study.
SNPs in DNMTs genes have primarily been investigated as risk factors for cancer susceptibility given that aberrant DNA methylation is one of the earliest molecular events during carcinogenesis [22]. In addition, since SNPs in DNMTs genes influence DNA methylation at global and gene-specific levels, they have also been also investigated as susceptibility factors for common complex diseases other than cancer [23]. Among the SNPs investigated in the present study, DNMT3A rs1550117 and DNMT3B rs2424908 have been associated with a lower cancer risk, as shown by recent meta-analyses [24,25]. With regard to DNMT1 rs2228611, the DNMT1 rs2228611GG genotype has been reported to be associated with a higher risk of ovarian cancer [26]. Although DNMT1 rs2228611 is a synonymous SNP located in exon 17, it has a possible splice regulatory function with A>G resulting in gaining of three exonic splicing enhancer binding motifs [23]. In addition, DNMT1 rs2228611 has significant modifying effects on the inverse relationship between LINE-1 methylation, a marker of global methylation, and cadmium exposure in Argentinean women [27], suggesting a possible role in regulating the methylation level in response to environmental cues.
Reactive oxygen species (ROS) caused by radiation could be a common trigger mechanism in both fibrosis and radiation response. In oxygenated tumor and normal cells, mitochondria play a major role in the generation of ROS and appear to be the primary loci of RT effects [14]. Conversely, mitochondrial polymorphisms defining major haplogroups have been suggested to alter oxidative phosphorylation coupling as well as ROS production [28]. In addition, cybrids containing mitochondrial haplogroup H have lower levels of DNMT1, DNMT3A, and DNMT3B and global methylation than those with mitochondrial haplogroup J [15]. Based on our previous findings highlighting a protective role of mitochondrial haplogroup H on radiation-induced fibrosis [18], we built a GRS based on its combination with DNMT1 rs2228611. In line with the concern recently raised for high risk of over-fitting and false positive results of multifactorial genetic models [20], for the GRS construction we only considered DNMT SNPs significantly associated with radiationinduced fibrosis, rather than using the total number of putative risk alleles. Our results provided evidence of an increased discrimination accuracy when DNMT1 rs2228611 was combined with mitochondrial haplogroup H, suggesting potential usefulness of rs2228611 in genetic-based models for prediction of radiation-induced skin fibrosis. In addition, the method used for weighting predictors, which was based on Cox hazard regression coefficients, can be easily applied for inclusion of clinical factors as well. Unfortunately, none of the clinical or dosimetric variables analyzed emerged as independent predictors for grade ≥ 2 fibrosis, as we reported in our previous analysis [18].
Our findings should be interpreted in light of the following limitations and considerations. First, the lack of a second cohort of breast cancer patients precluded the possibility of validating DNMT1 rs2228611 as a predictive factor for radiation-induced fibrosis. Thus, our results need to be interpreted cautiously because of the small sample size and risk of false positive findings given that very few radiogenomic studies with a candidate gene approach have been replicated in independent cohorts [17]. Second, in the present study we did not evaluate gene polymorphisms involved in enzymatic DNA demethylation, such as the ten-eleven translocation (TET) protein family, which are known to play a role in fibrogenesis [29]. Therefore, a more comprehensive investigation of genes encoding epigenetic enzymes should be conducted to develop and validate a clinically useful geneticbased model for prediction of radiation-induced skin fibrosis. Third, the predictive performance of the genetic model, based solely on rs2228611 and haplogroup H, is rather modest. However, this is expected for complex polygenic traits such as clinical radiosensitivity, which is probably the result of the combined effects of multiple polymorphic loci [7]. In addition, further investigation in cellular and in vivo animal models is still required to provide mechanistic insights into the protective role of rs2228611GG on radiation-induced fibrosis.
ConclusionOur findings support a protective effect of the DNMT1 rs2228611GG genotype on the risk of radiation-induced fibrosis in breast cancer patients. In addition, we provide evidence that inclusion of DNMT1 rs2228611 in a genetic-based model has the potential to increase prediction accuracy for radiation-induced fibrosis. However, further investigation to provide mechanistic insights into the role of DNMT1 rs2228611 in fibrogenesis and radiation response is warranted. In addition, a more comprehensive investigation of the role of germline polymorphisms should be conducted to develop and validate a clinically useful genetic-based model for prediction of radiation-induced fibrosis after treatment of breast cancer.
AcknowledgmentsThis work was funded by grants from Fondazione della Comunità del Novarese, by Regione Piemonte, Ricerca Sanitaria Finalizzata to AAG (2006) and to PLC (2003, 2007 and 2008). The “Lega Italiana per la lotta contro i tumori”, Section of Vercelli, Italy, supported LD.
Fig. 1.Kaplan-Meier plot of cumulative grade ≥ 2 radiation-induced fibrosis in breast cancer patients by DNMT1 rs2228611 genotypes (p=0.018, log-rank test). 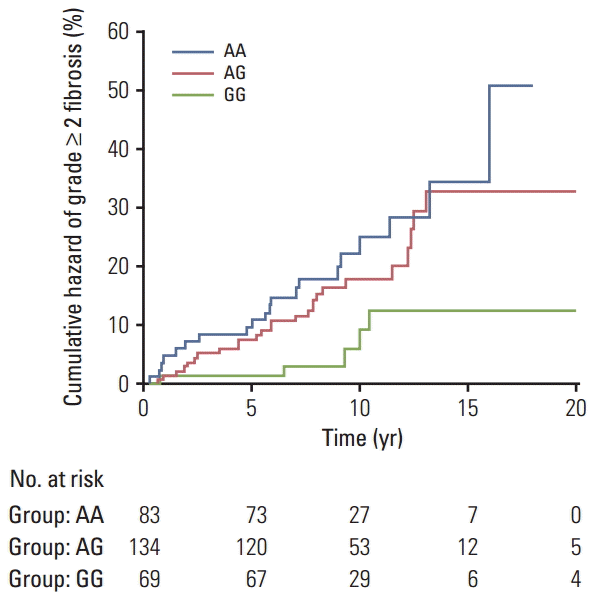
Fig. 2.Receiver operating characteristic curve curves for prediction of grade ≥ 2 radiation-induced fibrosis based on different combinations of DNMT1 rs2228611 and/or mitochondrial haplogroup H, as described in Table 3. Model 5 vs. model 1, p=0.053; model 5 vs. model 3, p=0.009; model 5 vs. model 4, p=0.93. Model 2 is not shown since it is equivalent to model 3. AUC, area under the receiver operating characteristic curves; CI, confidence interval. 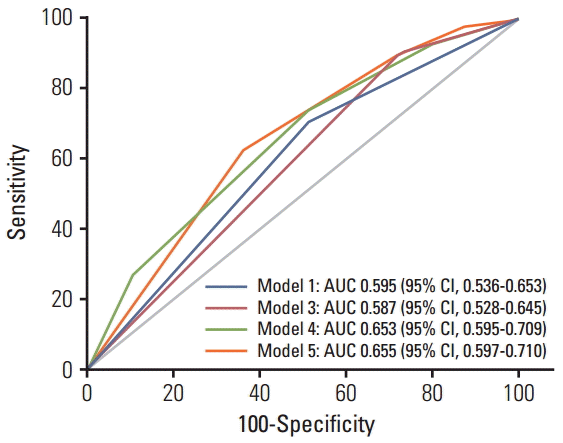
Fig. 3.Kaplan-Meier plot of cumulative grade ≥ 2 radiation-induced fibrosis in breast cancer patients by sum risk scores based on model 5, which combines mitochondrial haplogroup H with the recessive contrast of DNMT1 rs2228611 (p trend=0.0005, log-rank test). 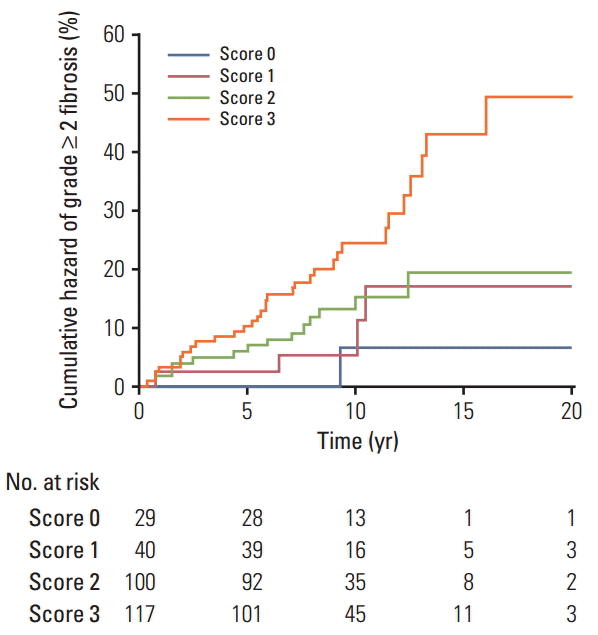
Table 1.Association analysis between polymorphisms of DNA methyltransferase genes and risk of grade ≥ 2 radiation-induced fibrosis (LENT-SOMA scale) in breast cancer patients
Table 2.Association analysis between XRCC1 rs2682585 and risk of grade ≥ 2 radiation-induced fibrosis (LENT-SOMA scale) in breast cancer patients
Table 3.Coefficients and risk points of each genetic predictor for grade ≥ 2 fibrosis in Cox regression models References1. Smith IE, Ross GM. Breast radiotherapy after lumpectomy: no longer always necessary. N Engl J Med. 2004;351:1021–3.
2. O’Sullivan B, Levin W. Late radiation-related fibrosis: pathogenesis, manifestations, and current management. Semin Radiat Oncol. 2003;13:274–89.
3. Barnett GC, West CM, Dunning AM, Elliott RM, Coles CE, Pharoah PD, et al. Normal tissue reactions to radiotherapy: towards tailoring treatment dose by genotype. Nat Rev Cancer. 2009;9:134–42.
4. Deantonio L, Gambaro G, Beldi D, Masini L, Tunesi S, Magnani C, et al. Hypofractionated radiotherapy after conservative surgery for breast cancer: analysis of acute and late toxicity. Radiat Oncol. 2010;5:112.
5. Lilla C, Ambrosone CB, Kropp S, Helmbold I, Schmezer P, von Fournier D, et al. Predictive factors for late normal tissue complications following radiotherapy for breast cancer. Breast Cancer Res Treat. 2007;106:143–50.
6. Popanda O, Marquardt JU, Chang-Claude J, Schmezer P. Genetic variation in normal tissue toxicity induced by ionizing radiation. Mutat Res. 2009;667:58–69.
7. Barnett GC, Thompson D, Fachal L, Kerns S, Talbot C, Elliott RM, et al. A genome wide association study (GWAS) providing evidence of an association between common genetic variants and late radiotherapy toxicity. Radiother Oncol. 2014;111:178–85.
9. Hackett JA, Surani MA. DNA methylation dynamics during the mammalian life cycle. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110328.
10. Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, TontiFilippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22.
11. Neary R, Watson CJ, Baugh JA. Epigenetics and the overhealing wound: the role of DNA methylation in fibrosis. Fibrogenesis Tissue Repair. 2015;8:18.
12. Weigel C, Schmezer P, Plass C, Popanda O. Epigenetics in radiation-induced fibrosis. Oncogene. 2015;34:2145–55.
13. Antwih DA, Gabbara KM, Lancaster WD, Ruden DM, Zielske SP. Radiation-induced epigenetic DNA methylation modification of radiation-response pathways. Epigenetics. 2013;8:839–48.
14. Richardson RB, Harper ME. Mitochondrial stress controls the radiosensitivity of the oxygen effect: implications for radiotherapy. Oncotarget. 2016;7:21469–83.
15. Atilano SR, Malik D, Chwa M, Caceres-Del-Carpio J, Nesburn AB, Boyer DS, et al. Mitochondrial DNA variants can mediate methylation status of inflammation, angiogenesis and signaling genes. Hum Mol Genet. 2015;24:4491–503.
16. Bellizzi D, D'Aquila P, Giordano M, Montesanto A, Passarino G. Global DNA methylation levels are modulated by mitochondrial DNA variants. Epigenomics. 2012;4:17–27.
17. Seibold P, Behrens S, Schmezer P, Helmbold I, Barnett G, Coles C, et al. XRCC1 polymorphism associated with late toxicity after radiation therapy in breast cancer patients. Int J Radiat Oncol Biol Phys. 2015;92:1084–92.
18. Terrazzino S, Deantonio L, Cargnin S, Donis L, Pisani C, Masini L, et al. Common European mitochondrial haplogroups in the risk for radiation-induced subcutaneous fibrosis in breast cancer patients. Clin Oncol (R Coll Radiol). 2016;28:365–72.
19. Pavy JJ, Denekamp J, Letschert J, Littbrand B, Mornex F, Bernier J, et al. EORTC Late Effects Working Group. Late effects toxicity scoring: the SOMA scale. Int J Radiat Oncol Biol Phys. 1995;31:1043–7.
20. Andreassen CN. A simulated SNP experiment indicates a high risk of over-fitting and false positive results when a predictive multiple SNP model is established and tested within the same dataset. Radiother Oncol. 2015;114:310–3.
21. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45.
22. Pouliot MC, Labrie Y, Diorio C, Durocher F. The role of methylation in breast cancer susceptibility and treatment. Anticancer Res. 2015;35:4569–74.
23. Saradalekshmi KR, Neetha NV, Sathyan S, Nair IV, Nair CM, Banerjee M. DNA methyl transferase (DNMT) gene polymorphisms could be a primary event in epigenetic susceptibility to schizophrenia. PLoS One. 2014;9:e98182
24. Zhang W, Xu Y, Ma G, Qi W, Gu H, Jiang P. Genetic polymorphism of DNA methyltransferase 3A rs1550117 A>G and risk of cancer: a meta-analysis. J Invest Surg. 2015;28:346–53.
25. Duan F, Cui S, Song C, Dai L, Zhao X, Zhang X. Systematic evaluation of cancer risk associated with DNMT3B polymorphisms. J Cancer Res Clin Oncol. 2015;141:1205–20.
26. Mostowska A, Sajdak S, Pawlik P, Lianeri M, Jagodzinski PP. DNMT1, DNMT3A and DNMT3B gene variants in relation to ovarian cancer risk in the Polish population. Mol Biol Rep. 2013;40:4893–9.
27. Hossain MB, Vahter M, Concha G, Broberg K. Low-level environmental cadmium exposure is associated with DNA hypomethylation in Argentinean women. Environ Health Perspect. 2012;120:879–84.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||