AbstractPurposeHomeobox (HOX) genes are essential developmental regulators that should normally be in the silenced state in an adult brain. The aberrant expression of HOX genes has been associated with the prognosis of many cancer types, including glioblastoma (GBM). This study examined the identity and role of HOX genes affecting GBM prognosis and treatment resistance.
Materials and MethodsThe full series of HOX genes of five pairs of initial and recurrent human GBM samples were screened by microarray analysis to determine the most plausible candidate responsible for GBM prognosis. Another 20 newly diagnosed GBM samples were used for prognostic validation. In vitro experiments were performed to confirm the role of HOX in treatment resistance. Mediators involved in HOX gene regulation were searched using differentially expressed gene analysis, gene set enrichment tests, and network analysis.
ResultsThe underexpression of HOXA11 was identified as a consistent signature for a poor prognosis among the HOX genes. The overall survival of the GBM patients indicated a significantly favorable prognosis in patients with high HOXA11 expression (31±15.3 months) compared to the prognoses in thosewith low HOXA11 expression (18±7.3 months, p=0.03). When HOXA11 was suppressed in the GBM cell lines, the anticancer effect of radiotherapy and/or temozolomide declined. In addition, five candidate mediators (TGFBR2, CRIM1, TXNIP, DPYSL2, and CRMP1) that may confer an oncologic effect after HOXA11 suppression were identified.
IntroductionHomeobox (HOX) genes are essential developmental regulators that control a wide range of processes, including apoptosis, differentiation, motility, and angiogenesis [1]. In humans, there are four HOX clusters (A, B, C, and D), and 39 HOX genes have been identified [1]. The HOX genes are normally active during embryogenesis, but most are not expressed or are expressed at very low levels in an adult brain [2]. On the other hand, several studies have indicated aberrant expression of the HOX genes in brain tumors as well in as other cancers from various organs [3-6]. Moreover, there is growing clinical evidence of a prognostic effect of HOX gene expression in several cancers [7-10].
Aberrantly expressed HOX genes in cancer cells have multicapacity functions, including metastasis, tumor growth, anti-apoptosis, and differentiation suppression [1]. The overexpression of multiple HOX genes have been observed in glioblastoma (GBM) cell lines and primary astrocytoma [4]. In addition, some studies have shown that the HOX genes are important in the treatment resistance of GBM [11-13]. On the other hand, the precise mechanism showing the role of HOX genes and their functional relevance in glioma cells is unclear. GBMs, similar to other cancers, harbor a cell subpopulation with a stem cell-like capacity that is associated with the development of a tumor progeny and treatment resistance [9,14,15]. Given the roles of HOX genes in development and organogenesis, it has been postulated that a portion of the relative expression of HOX genes is integral to stem cell activity, specifically self-renewal, tissue specificity, and quiescence [1]. Among the HOX genes, the HOXA cluster is important in human embryonic stem cell differentiation [16]. Collectively, these results suggest that HOX genes are plausible candidates as biomarkers for assessing the prognosis of GBM and a credible target for overcoming the treatment resistance in GBM.
HOX genes were previously reported to be the genes of interest related to GBM recurrence and treatment resistance [17]. Moreover, a mechanism through which the HOXA10 gene affects temozolomide (TMZ) resistance in GBM cell lines was reported [12]. HOXA10 induces the transcription of early growth response 1, which results in phosphatase and tensin homolong (PTEN) and Rad51 paralogs. As a result, the homologous recombination DNA repair system with Rad51 genes can protect cancer cells from TMZ-induced cytotoxicity [12].
In the present study, this research was extended to another HOX gene, HOXA11, and its role in GBM prognosis was examined.
Materials and Methods1. Patient samples and cell linesFresh frozen tumor tissue samples of five GBM patients, in whom of pairs of initial and recurrent samples were available for screening, and another 20 newly diagnosed GBM patients for validation were used in this study. All patients were managed using a standard GBM treatment protocol of concurrent radiotherapy and TMZ treatment, followed by adjuvant TMZ as a primary treatment. The tumor tissues were obtained during surgery, snap-frozen in liquid nitrogen, and stored at −80°C prior to use. The study was approved by an institutional review committee.
The human glioma U251, U373, and LN18 cell lines were purchased from the American Type Culture Collection (Manassas, VA), and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 5% antibiotics (streptomycin) in a humidified atmosphere containing 5% CO2 and 95% air at 37°C.
2. Reverse transcription polymerase chain reactionThe cell lines were lysed with TRIzol (Life Technologies, Carlsbad, CA), and RNA isolation was performed using an RNeasy Mini Kit (#74104, Qiagen, Valencia, CA). The total RNA was treated with DNase and then quantified by spectrophotometry. In addition, cDNA was synthesized from 1 μg of the total RNA using a reverse transcription kit (#205311, Qiagen) according to the manufacturer’s protocol. The primers used were designed using an online primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primer sequences for HOXA11 were 5'-GATTTCTCCAGCCTCCCTTC-3' (forward) and 5'-AGAAATTGGACGAGACTGCG-3' (reverse). Using these primers, a reverse transcription polymerase chain reaction (RT-PCR) was performed for 35 cycles. Each cycle was comprised of 95°C for 30 seconds, 62°C for 30 seconds, and 72°C for 45 seconds with each primer set. The RT-PCR products were resolved by 2% agarose gel electrophoresis.
3. Western blotThe whole protein extracts from the tissue samples were prepared using a PRO-PREP lysis buffer (#17081, iNtRon Biotechnology, Seongnam, Korea), and the protein concentrations were determined using a bicinchoninic acid protein assay (#23227, Thermo Fisher Scientific, Waltham, MA). The proteins were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, followed by blotting onto nitrocellulose membranes, and probing with the antibodies against HOXA11 (#SC-48542, 1:500 dilution, Santa Cruz Biotechnology, Santa Cruz, CA). The blotted membranes were then incubated with the goat anti-rabbit IgG secondary antibody for 1 hour. Subsequently, the membranes were incubated in an Amersham ECL-prime solution (#RPN2232, GE Healthcare Life Sciences, Pittsburgh, PA) in the dark for 1 minute and exposed under FluorChemHD2 (Cell Biosciences, Santa Clara, CA) for visualization. Only the samples with a consistent result from repeated experiments were selected for analysis. The densities of the bands were measured using free image analyzer software (ImageJ V1.8x, National Institutes of Health, http://rsb.info.nih.gov/ij/). The results are presented as the mean±standard error of mean calculated from independent samples.
4. RNA interferenceTo knock down HOXA11 expression in cells, small interfering RNA (siRNA) experiments were performed using commercially available sequences targeting HOXA11 (#SASI-Hs01-00110410, #SASI-Hs01-00110413, and #SASI-Hs01-00110417, Sigma Aldrich, St. Louis, MO) as well as with the non-targeting control siRNA (#D-001610-01-05, Dharmacon, Lafayette, CO). When the cells reached 70%-80% confluence, they were transfected with siRNA under the most efficient transfection condition, as determined by the NEON Transfection system (#MPK5000, Life Technologies). The cells were cultured in media without antibiotics to increase the siRNA transfection efficiency for 24 hours.
5. Cell viability analysis after drug and radiation treatmentThe control and transfected cells were grown on 96-well plates at a density of 4×103 cells per well for 24 hours. Subsequently, the cells were either treated with TMZ (#ALX-420-044-M100, Enzo Life Sciences, Farmingdale, NY) to a final concentration of 300 μg/mL for 24 hours or irradiated with 4 MV X-rays from a linear accelerator (Clinac 4/100, Varian Medical Systems, Palo Alto, CA) at a dose rate of 10.0 Gy/min. For the combination treatment, the cells were irradiated first and then treated with TMZ.
Cell viability analysis was performed using a Colorimetric Cell Counting Kit-8 (CCK; Dojindo Molecular Technologies, Kumamoto, Japan). The number of viable cells was quantified according to the manufacturer’s instructions by reading the ultraviolet absorption spectra at 450 nm on a microplate 2 hours after adding 10 μL of a CCK solution per well. All experiments were conducted in triplicate.
6. MicroarrayFor tissue sample analysis, the total RNA was extracted from the tissue samples using the mirVana miRNA Isolation Kit (#AM1560, Ambion, Austin, TX) for microarray analysis after quantification and qualification. The total RNA quality was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The cut off RNA integrity number for RNA used in RNA amplification was 7.0 or above. The cRNA was produced using an Illumina TotalPrep RNA Amplification Kit (#IL1791, Ambion) according to the provided protocol. The cRNA was used for hybridization to a human HT12-v4 Illumina Beadchip gene expression array (Illumina, San Diego, CA) according to the manufacturer’s protocol. The arrays were scanned and the fluorescence signals were obtained using an Illumina BeadArray Reader (BeadStation 500GXDW, Illumina). The signal obtained from the scanned beadchip was transformed to intensity raw data using GenomeSortudio software (ver. 2009.1, Illumina) and was used for further data analysis. The raw data were normalized by applying a log2 transformation, quantile normalization, and gene and array centering. All data processing was performed using the R/Bioconductor packages (ver. 2.14, http://www.bioconductor.org).
To determine the changes in gene expression before and after HOXA11 knockdown, the total RNA extracted from the LN18 cells transduced with siHOXA11 or control siRNA were analyzed by Affymetrix GeneChip Human Gene 1.0ST Arrays (Affymetrix, Santa Clara, CA). The RNA was amplified and labeled using a GeneChip WT Sense Target Labeling and Control Reagents Kit (Affymetrix). The cDNA was synthesized, labeled, and hybridized to the GeneChip array according to the manufacturer's protocol. The GeneChips were washed and stained using the GeneChip Fluidics Station 450 (Affymetrix), and then scanned using a GeneChip Scanner 3000 7G (Affymetrix). The expression data were normalized using the robust multi-array average method. Affymetrix Expression Console ver. 1.1 (Affymetrix) was used to compare the group signals, and the data were logtransformed (base 2) for parametric analysis. The differentially expressed genes (DEGs) were identified by significance analysis of the microarrays method in the R package ‘samr’ (R 2.11.1).
7. Statistical analysisThe data from the experiments were tested for their significance using an unpaired two-tailed Student's t test. An ANOVA and Student's t test were used to identify the significant differences in cell death rates. Kaplan-Meier curve analysis was used to generate the overall survival curves. The differences between the survival curves were analyzed using a log-rank test. The results were analyzed using IBM SPSS Statistics software ver. 19.0 (IBM Co., Armonk, NY). The data are presented as mean±standard deviation for three or more separate experiments. A p-value of 0.05 or lower was considered significant.
For microarray analyses, the false discovery rates (FDRs) were calculated using three GenePattern software modules (http://www.broadinstitute.org/cancer/software/genepattern; ComparativeMarkerSelection ver. 10, HierarchicalClustering ver. 6, and HeatMapViewer ver. 13) [18]. The cutoff value for FDR significance was < 0.05. The significantly regulated genes were subjected to functional gene classification using the DAVID Bioinformatics Resources annotation tool (ver. 6.7, http://david.abcc.ncifcrf.gov/) [19]. The selected gene IDs of the identified DEGs were entered into GeneMANIA software (ver. 3.1.2.8, http://www.genemania.org) for network analysis [20].
Results1. Down-regulation of HOXA11 is associated with a poor prognosis in GBM patientsThe relative changes in HOX gene expression in five pairs of primary and recurrent GBM samples were assessed using microarray analysis (Fig. 1). Among the 39 HOX genes, HOXA11 was the only gene that consistently showed a significant down-regulation in the recurrent samples (p=0.046). The overall survival of a separate set of 20 GBM patients indicated significantly favorable prognoses in those with high HOXA11 expression compared to those with low HOXA11 protein expression (survival, 31±15.3 months with high HOXA11 expression vs. 18±7.3 months with low HOXA11 expression, p=0.03; expression status determined by western blot analysis) (Fig. 2). Therefore, the down-regulation of HOXA11 is associated with a poor prognosis in GBM patients.
2. Suppression of HOXA11 mediates the treatment resistance in vitroTo confirm the effects of HOXA11 expression on the resistance to the current standard treatment protocol for GBM, three malignant glioma cell lines (U251, U373, and LN18) were transduced with HOXA11 siRNA to assess the cell viability. The viable cell fractions were analyzed 72 hours after treatment with either single or combination applications of radiation treatment (RT) and TMZ. A significant increase in the number of cells with HOXA11 suppression after either RT or TMZ was observed (Table 1, Fig. 3). When HOXA11 was suppressed, the treatment resistance effect was more pronounced after treatment with a combination of RT and TMZ (p=0.022 for RT vs. RT/TMZ and p=0.053 for TMZ vs. RT/TMZ).
3. Mediators of HOXA11-related oncologic effectTo identify the genes under the regulation of HOXA11, the microarray expression profiling data of the control LN18 cells was compared with that of the HOXA11-silenced cells with HOXA11 siRNA. LN18 was chosen because it showed the most significant decrease in HOXA11 expression after siRNA transduction among the three cell lines tested. After normalization of the values and DEG analysis, 11 up-regulated and 51 down-regulated genes that exhibited more than two-fold changes after HOXA11 suppression were identified (Fig. 4, Supplementary dataset).
The functional annotation tools within DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/home.jsp) was then used for gene annotation enrichment analysis for the identified DEGs [19,21]. Three gene ontology categories (GO:0040008 regulation of growth, GO:0005739 mitochondrion, and GO:0004157 dihydropyrimidinase activity) were enriched significantly (EASE score < 0.05) in 11 genes (EI24, CRIM1, MUL1, TGFBR2, TXNIP, STXBP1, ELK3, COX7A2, NRAS, DPYSL2, and CRMP1) among the DEG set (Table 2). For deeper insight into the HOXA11 regulatory mechanism, network analysis was performed using GeneMANIA software (ver. 3.1.2.8) [20]. The network was constructed with the coexpression relationships, which revealed transforming growth factor, beta receptor 2 (TGFBR2), cysteine rich transmembrane BMP regulator 1 (CRIM1), thioredoxin interacting protein (TXNIP), dihydropyrimidinase-like 2 (DPYSL2), and collapsin response mediator protein 1 (CRMP1) genes to be the key hub regulators associated with HOXA11 suppression (Fig. 5).
DiscussionHOX genes, which are a cluster of master regulators of embryogenesis, are expressed temporarily during the developmental phase in vertebrates, but they should be silenced in the adult central nervous system [2,21,22]. Accumulating evidence of the aberrant expression of HOX genes in cancers suggests that these genes have diverse roles in oncogenesis [2,3,21-26]. In addition, a relationship between the HOX genes and treatment resistance or prognosis in cancer has frequently been proposed [9,11,12,27,28]. Several studies have revealed the tumor suppressor roles of HOX genes in many cancers [1]. Moreover, there is evidence suggesting that the restored expression of tumor suppressor HOX genes can attenuate the cancer progression in vitro and in vivo [29-31]. Experimental evidence of an association between HOXA10 and TMZ resistance in GBM has been presented [12]. Subsequent reports into the oncogenic role of HOXA10 in various cancers have been published [32-40]. On the other hand, based on the previous experimental data it is suspected that HOXA11 acts as a tumor suppressor in opposition to HOXA10, which prompted a further study of the function of HOXA11 in GBM.
The present study proposes a tumor suppressor function of HOXA11 in GBM based on the results from both in vitro experiments and human samples. The role HOXA11 in diverse cancers has been reported [41-47]. In gastric cancer, the epigenetic down-regulation of HOXA11 has been related to carcinogenesis, proliferation, migration, and invasion [41,42]. Similarly, in lung and ovarian cancers, the down-regulation of HOXA11 was shown to be a poor prognostic factor [43,44]. In GBM, the epigenetic down-regulation rate of HOXA11 was reported to be 51%-75%, and HOXA11 was one of the most frequently methylated genes in GBM [45-47]. The methylation of HOXA11 was associated with older patient ages and poor survival in GBM [47]. In addition to the evidence of the epigenetic characteristics of HOXA11 as a prognostic marker, the present study provides direct evidence of the prognostic value of HOXA11 expression in GBM samples (Figs. 1 and 2). Based on previous reports and the results in the present study, it is obvious that HOXA11 is a tumor suppressor in GBM and other cancers.
Treatment resistance induced by HOXA11 down-regulation, as detected by in vitro experiments in this study, is a mechanism contributing to a poor prognosis. In addition, this study detected candidate mediators (TGFBR2, CRIM1, TXNIP, DPYSL2, and CRMP1) that may impart treatment resistance after HOXA11 suppression. HOXA11 was reported to be a tumor suppressor gene that can sensitize a chemotherapeutic agent in ovarian cancer [48,49]. On the other hand, there is little direct evidence of treatment resistance induced by HOXA11 suppression in other cancer types. Nevertheless, these results provide a promising basis for the development of HOXA11 applications that target chemo- or radio-sensitizers in GBM. Among the five candidate mediators of HOXA11 suppression-induced oncologic effect, none have been reported to be associated with a HOXA11 regulatory mechanism. On the other hand, CRMP1 is notable because it is an invasion-suppressor gene in cancer cells [50]. Recently, it was suggested that decreased CRMP1 expression in GBMs harboring EGFRvIII positivity is responsible for promoting invasion [51]. Moreover, those authors proposed the counter-activation of Rac-1 after CRMP1 suppression as a mechanistic hypothesis for the invasive phenotype [51]. Other evidence regarding the role of HOXA11 in oncogenesis includes a report on the regulation of matrix metalloproteinase 2 expression, which can affect cancer cell migration and invasion [52]. Overall, this study postulates an oncogenic pathway involving EGFRvIII, HOXA11, CRMP1, and Rac-1 in GBM, which will require further investigation.
Electronic Supplementary MaterialSupplementary materials are available at Cancer Research and Treatment website (http://www.e-crt.org).
AcknowledgmentsThis research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2015-R1D1A1A09057171) in Korea and Seoul National University Hospital Research Fund (04-2010-0470).
Fig. 1.Relative changes in the expression of homeobox (HOX) genes among five pairs of primary and recurrent glioblastoma (GBM) samples as determined by microarray analysis. (A) Heatmap and hierarchical clustering analysis showed inconsistent results in sequential HOX gene expression changes between the primary and recurrent samples, except for HOXA11. (B) HOXA11 gene is the only HOX gene that was consistently down-regulated in the recurrent GBM samples compared to primary samples for all five sample pairs. 
Fig. 2.(A) Results of western blot analysis for normalized HOXA11 expression. The patients were grouped according to the HOXA11 expression macroscopically, and the difference in expression level was confirmed by intensity measurement of the bands. (B) The overall survival of a separate cohort of glioblastoma patients by HOXA11 expression. The survival was significantly longer in patients with high HOXA11 expression (31±15.3 months) than in those with low HOXA11 expression (18±7.3 months, p=0.037). 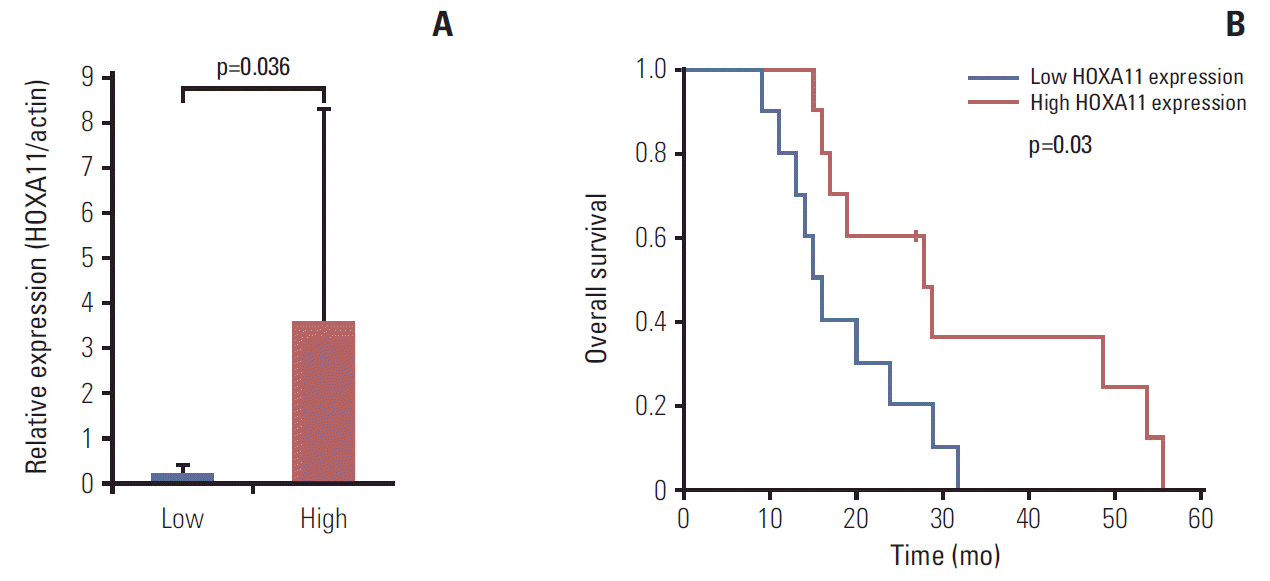
Fig. 3.Treatment resistance of glioma cell lines (U251, U373, and LN18) assessed by cell viability tests after the inhibition of HOXA11 and presented as relative viable cell fractions. Direct inhibition of HOXA11 by small interfering RNA (siRNA) results in a significant increase in cell survival, 72 hours after single or combination applications of radiation (RT) and temozolomide (TMZ) treatments (*p < 0.05). 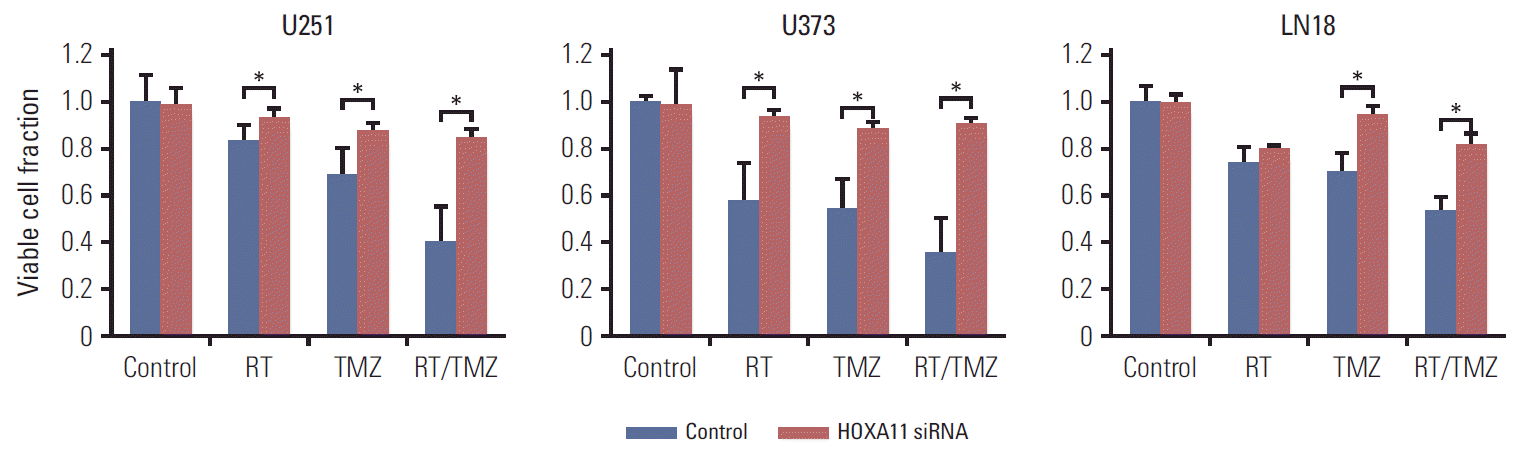
Fig. 4.Identification of the HOXA11-regulated genes in LN18 cells. (A) Reverse transcription polymerase chain reaction results from LN18 cells after HOXA11 knockdown by siRNA. (B) HOXA11-silencing siRNA versus control siRNA transduced cells compared using Affymetrix GeneChip Human Gene 1.0ST Arrays. The probe sets with fold changes greater than 2-fold are shown. After HOXA11 suppression, 11 up-regulated and 51 down-regulated genes were identified. (C) MA plots showing the distribution of regulated genes. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IQR, interquartile range; siRNA, small interfering RNA. 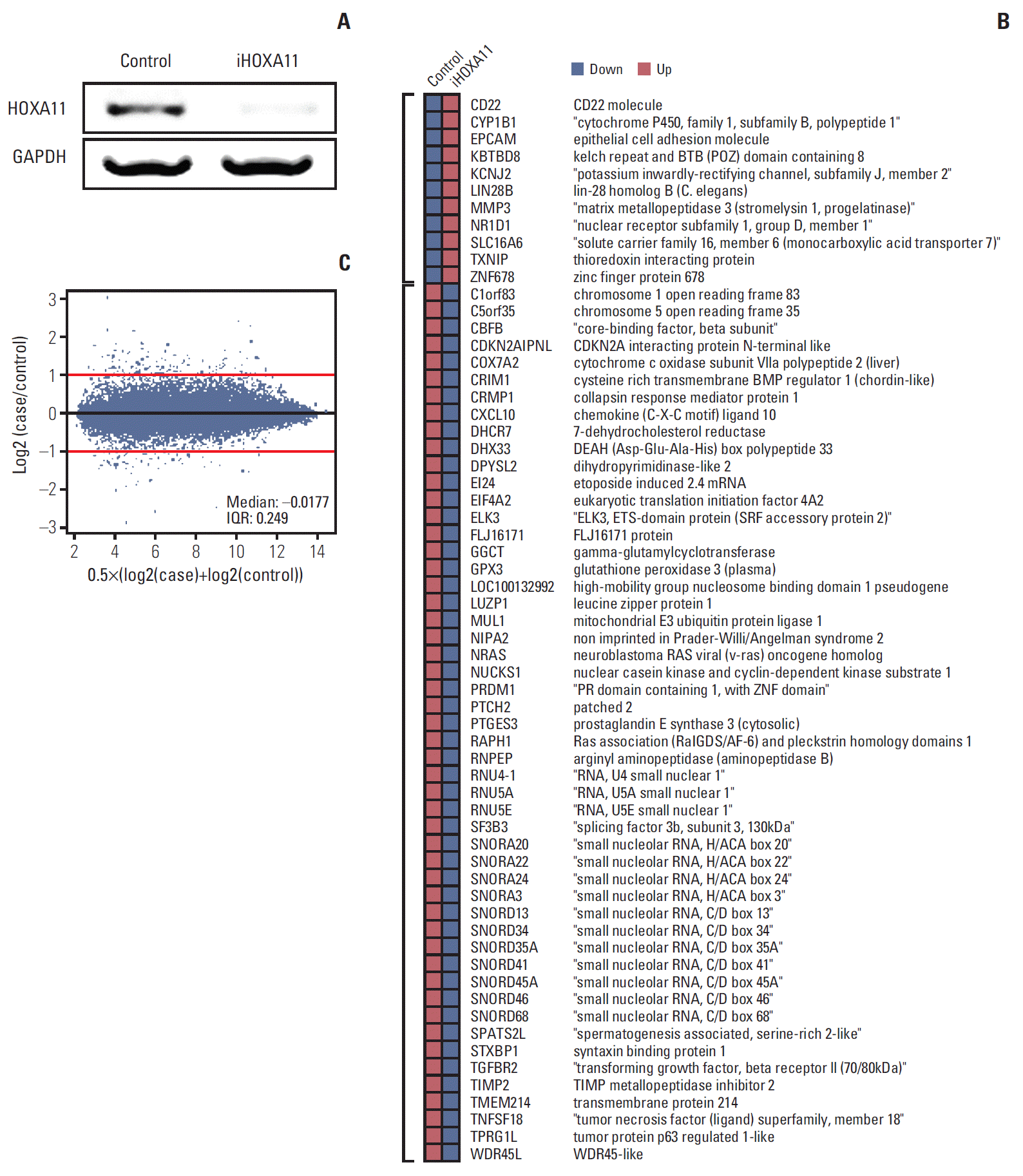
Fig. 5.Coexpression network incorporating selected gene sets drawn from the functional gene annotation enrichment analysis of differentially expressed genes after HOXA11 suppression. The network was constructed based on coexpression interactions using GeneMANIA software (ver. 3.1.2.8). The key hub regulators associated with HOXA11 suppression are expressed as black solid circles. 
Table 1.Differences in the survival fraction after the direct inhibition of HOXA11 by siRNA compared with the control in cells 72 hours after treatment with single or combination applications of RT and TMZ treatments
Table 2.Enriched gene ontology categories in the genes expressed differently after HOXA11 suppression in LN18 cells References2. Takahashi Y, Hamada J, Murakawa K, Takada M, Tada M, Nogami I, et al. Expression profiles of 39 HOX genes in normal human adult organs and anaplastic thyroid cancer cell lines by quantitative real-time RT-PCR system. Exp Cell Res. 2004;293:144–53.
3. Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–85.
4. Abdel-Fattah R, Xiao A, Bomgardner D, Pease CS, Lopes MB, Hussaini IM. Differential expression of HOX genes in neoplastic and non-neoplastic human astrocytes. J Pathol. 2006;209:15–24.
5. Bodey B, Bodey B Jr, Siegel SE, Kaiser HE. Immunocytochemical detection of the homeobox B3, B4, and C6 gene products in childhood medulloblastomas/primitive neuroectodermal tumors. Anticancer Res. 2000;20:1769–80.
6. Buccoliero AM, Castiglione F, Rossi Degl'Innocenti D, Ammanati F, Giordano F, Sanzo M, et al. Hox-D genes expression in pediatric low-grade gliomas: real-time-PCR study. Cell Mol Neurobiol. 2009;29:1–6.
7. Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102:262–8.
8. Miao J, Wang Z, Provencher H, Muir B, Dahiya S, Carney E, et al. HOXB13 promotes ovarian cancer progression. Proc Natl Acad Sci U S A. 2007;104:17093–8.
9. Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, et al. Stem cell-related "self-renewal" signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–24.
10. Wang Z, Dahiya S, Provencher H, Muir B, Carney E, Coser K, et al. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clin Cancer Res. 2007;13:6327–34.
11. Gaspar N, Marshall L, Perryman L, Bax DA, Little SE, Viana-Pereira M, et al. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Res. 2010;70:9243–52.
12. Kim JW, Kim JY, Kim JE, Kim SK, Chung HT, Park CK. HOXA10 is associated with temozolomide resistance through regulation of the homologous recombinant DNA repair pathway in glioblastoma cell lines. Genes Cancer. 2014;5:165–74.
13. Kim KJ, Moon SM, Kim SA, Kang KW, Yoon JH, Ahn SG. Transcriptional regulation of MDR-1 by HOXC6 in multidrug-resistant cells. Oncogene. 2013;32:3339–49.
14. Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60.
15. Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6.
16. Atkinson SP, Koch CM, Clelland GK, Willcox S, Fowler JC, Stewart R, et al. Epigenetic marking prepares the human HOXA cluster for activation during differentiation of pluripotent cells. Stem Cells. 2008;26:1174–85.
17. Kwon SM, Kang SH, Park CK, Jung S, Park ES, Lee JS, et al. Recurrent glioblastomas reveal molecular subtypes associated with mechanistic implications of drug-resistance. PLoS One. 2015;10:e0140528
18. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–1.
19. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
20. Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–20.
21. Hueber SD, Lohmann I. Shaping segments: Hox gene function in the genomic age. Bioessays. 2008;30:965–79.
22. Ohgo S, Itoh A, Suzuki M, Satoh A, Yokoyama H, Tamura K. Analysis of hoxa11 and hoxa13 expression during patternless limb regeneration in Xenopus. Dev Biol. 2010;338:148–57.
23. Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–76.
24. Bahrani-Mostafavi Z, Tickle TL, Zhang J, Bennett KE, Vachris JC, Spencer MD, et al. Correlation analysis of HOX, ErbB and IGFBP family gene expression in ovarian cancer. Cancer Invest. 2008;26:990–8.
25. Cillo C, Barba P, Freschi G, Bucciarelli G, Magli MC, Boncinelli E. HOX gene expression in normal and neoplastic human kidney. Int J Cancer. 1992;51:892–7.
26. De Vita G, Barba P, Odartchenko N, Givel JC, Freschi G, Bucciarelli G, et al. Expression of homeobox-containing genes in primary and metastatic colorectal cancer. Eur J Cancer. 1993;29A:887–93.
27. Costa BM, Smith JS, Chen Y, Chen J, Phillips HS, Aldape KD, et al. Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res. 2010;70:453–62.
28. Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW, Choi SH, et al. Overexpression of miR-196b and HOXA10 characterize a poor-prognosis gastric cancer subtype. World J Gastroenterol. 2013;19:7078–88.
29. Carrio M, Arderiu G, Myers C, Boudreau NJ. Homeobox D10 induces phenotypic reversion of breast tumor cells in a three-dimensional culture model. Cancer Res. 2005;65:7177–85.
30. Petrini M, Felicetti F, Bottero L, Errico MC, Morsilli O, Boe A, et al. HOXB1 restored expression promotes apoptosis and differentiation in the HL60 leukemic cell line. Cancer Cell Int. 2013;13:101.
31. Rhoads K, Arderiu G, Charboneau A, Hansen SL, Hoffman W, Boudreau N. A role for Hox A5 in regulating angiogenesis and vascular patterning. Lymphat Res Biol. 2005;3:240–52.
32. Carrera M, Bitu CC, de Oliveira CE, Cervigne NK, Graner E, Manninen A, et al. HOXA10 controls proliferation, migration and invasion in oral squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:3613–23.
33. Cui XP, Qin CK, Zhang ZH, Su ZX, Liu X, Wang SK, et al. HOXA10 promotes cell invasion and MMP-3 expression via TGFbeta2-mediated activation of the p38 MAPK pathway in pancreatic cancer cells. Dig Dis Sci. 2014;59:1442–51.
34. Kurscheid S, Bady P, Sciuscio D, Samarzija I, Shay T, Vassallo I, et al. Chromosome 7 gain and DNA hypermethylation at the HOXA10 locus are associated with expression of a stem cell related HOX-signature in glioblastoma. Genome Biol. 2015;16:16.
35. Li B, Cao X, Weng C, Wu Y, Fang X, Zhang X, et al. HoxA10 induces proliferation in human prostate carcinoma PC-3 cell line. Cell Biochem Biophys. 2014;70:1363–8.
36. Liborio-Kimura TN, Jung HM, Chan EK. miR-494 represses HOXA10 expression and inhibits cell proliferation in oral cancer. Oral Oncol. 2015;51:151–7.
37. Oue N, Sentani K, Sakamoto N, Yasui W. Clinicopathologic and molecular characteristics of gastric cancer showing gastric and intestinal mucin phenotype. Cancer Sci. 2015;106:951–8.
38. Xiao ZD, Jiao CY, Huang HT, He LJ, Zhao JJ, Lu ZY, et al. miR-218 modulate hepatocellular carcinoma cell proliferation through PTEN/AKT/PI3K pathway and HoxA10. Int J Clin Exp Pathol. 2014;7:4039–44.
39. Zhang HY, Li JH, Li G, Wang SR. Activation of ARK5/miR-1181/HOXA10 axis promotes epithelial-mesenchymal transition in ovarian cancer. Oncol Rep. 2015;34:1193–202.
40. Zhang L, Wan Y, Jiang Y, Ma J, Liu J, Tang W, et al. Upregulation HOXA10 homeobox gene in endometrial cancer: role in cell cycle regulation. Med Oncol. 2014;31:52.
41. Bai Y, Fang N, Gu T, Kang Y, Wu J, Yang D, et al. HOXA11 gene is hypermethylation and aberrant expression in gastric cancer. Cancer Cell Int. 2014;14:79.
42. Cui Y, Gao D, Linghu E, Zhan Q, Chen R, Brock MV, et al. Epigenetic changes and functional study of HOXA11 in human gastric cancer. Epigenomics. 2015;7:201–13.
43. Fiegl H, Windbichler G, Mueller-Holzner E, Goebel G, Lechner M, Jacobs IJ, et al. HOXA11 DNA methylation: a novel prognostic biomarker in ovarian cancer. Int J Cancer. 2008;123:725–9.
44. Hwang JA, Lee BB, Kim Y, Park SE, Heo K, Hong SH, et al. HOXA11 hypermethylation is associated with progression of non-small cell lung cancer. Oncotarget. 2013;4:2317–25.
45. Laffaire J, Everhard S, Idbaih A, Criniere E, Marie Y, de Reynies A, et al. Methylation profiling identifies 2 groups of gliomas according to their tumorigenesis. Neuro Oncol. 2011;13:84–98.
46. Martinez R, Martin-Subero JI, Rohde V, Kirsch M, Alaminos M, Fernandez AF, et al. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics. 2009;4:255–64.
47. Skiriute D, Vaitkiene P, Asmoniene V, Steponaitis G, Deltuva VP, Tamasauskas A. Promoter methylation of AREG, HOXA11, hMLH1, NDRG2, NPTX2 and Tes genes in glioblastoma. J Neurooncol. 2013;113:441–9.
48. Fang F, Munck J, Tang J, Taverna P, Wang Y, Miller DF, et al. The novel, small-molecule DNA methylation inhibitor SGI-110 as an ovarian cancer chemosensitizer. Clin Cancer Res. 2014;20:6504–16.
49. Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72:2197–205.
50. Shih JY, Lee YC, Yang SC, Hong TM, Huang CY, Yang PC. Collapsin response mediator protein-1: a novel invasion-suppressor gene. Clin Exp Metastasis. 2003;20:69–76.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||