AbstractPurposeThe purpose of this study is to investigate the prognostic significance of SOX2 gene amplification and expression in patients with American Joint Committee on Cancer stage III lung squamous cell carcinoma (SCC) who underwent surgery followed by adjuvant radiotherapy.
Materials and MethodsPathological specimens were obtained from 33 patients with stage III lung SCC treated with surgery followed by adjuvant radiotherapy between 1996 and 2008. SOX2 gene amplification and protein expression were analyzed using fluorescent in situ hybridization and immunohistochemistry, respectively. Patients were divided into two groups according to their SOX2 gene amplification and protein expression status. Kaplan-Meier estimates and a Cox proportional hazards model were used to identify the prognostic factors affecting patient survival.
ResultsThe median follow-up period for surviving patients was 58 months (range, 5 to 102 months). SOX2 gene amplification was observed in 22 patients and protein overexpression in 26 patients. SOX2 overexpression showed significant association with SOX2 gene amplification (p=0.002). In multivariate analysis, SOX2 overexpression was a significant prognostic factor for overall survival (OS) (hazard ratios [HR], 0.1; 95% confidence interval [CI], 0.002 to 0.5; p=0.005) and disease-free survival (DFS) (HR, 0.15; 95% CI, 0.04 to 0.65; p=0.01). Age (HR, 0.33; 95% CI, 0.11 to 0.98; p=0.046) and total radiation dose (HR, 0.13; 95% CI, 0.02 to 0.7; p=0.02) were the independent prognostic factors for OS and DFS. Patients with SOX2 amplification did not show a longer OS (p=0.95) and DFS (p=0.48).
IntroductionLung cancer has one of the highest cancer mortality rates in several countries worldwide, including Korea [1,2]. Although cancer treatments have improved in recent decades, lung cancer remains non-responsive to curative treatments. However, novel emerging molecular techniques have made it possible to investigate its molecular-level pathophysiology, identify specific causative oncogenes or tumor suppressor genes, and make progress toward new paradigms for cancer treatment. Compared with other cancers, lung cancer has an increased number of well-defined genetic changes and/or abnormalities and more molecularly targeted agents have been applied in clinical practice for its treatment. However, most of the molecularly targeted agents used in clinical practice are for adenocarcinoma, and few are available for squamous cell carcinoma.
SOX2, a transcription factor encoded by the gene located at 3q26.33, plays a role in maintenance of embryonic stem cells. In addition, it is a “Yamanaka factor” which induces pluripotent stem cells from somatic cells, along with c-Myc, Oct4, and KLF4. It is also involved in morphogenesis and homeostasis of the esophageal, tracheobronchial, and bronchiolar epithelium [3].
SOX2 is expressed in bronchial epithelial cells, though not in alveolar cells or adenocarcinoma precursor lesions [4-6]. SOX2 is found exclusively in squamous cell carcinoma, where it is amplified and overexpressed at the gene and protein levels, respectively [3,6]. Previous studies have analyzed the prognosis of heterogeneously treated squamous cell carcinoma patient populations. However, only a small number of studies have investigated the association between SOX2 and the clinical outcome of patients treated with radiotherapy [7,8]. The aim of this study was to assess the prognostic significance of SOX2 amplification and expression in squamous cell lung cancer patients treated using adjuvant radiotherapy.
Materials and Methods1. Patient selectionThis retrospective study was approved by the Institutional Review Board of our institution (IRB No. 4-2012-0709). A total of 158 non-small cell lung cancer (NSCLC) patients who underwent pulmonary resection followed by adjuvant radiotherapy between 1996 and 2008 were identified. Squamous cell carcinoma was confirmed surgically in 71 patients, and clinical specimens were available from 36 patients. Of these, three patients were excluded from the analysis because of a lack of tumor tissue in their paraffin-embedded tissue blocks. Therefore, tumor samples from 33 patients were available for analysis. The patients’ medical records were reviewed retrospectively for evaluation of clinicopathological characteristics and survival outcomes. After radical resection, all patients were determined to have pathologic TNM stage III tumors according to the sixth edition cancer staging guidelines provided by the American Joint Committee on Cancer [9].
2. Fluorescent in situ hybridizationFluorescent in situ hybridization (FISH) was performed for analysis of SOX2 gene amplifications on 4-μm-thick formalin-fixed paraffin embedded (FFPE) tissue sections. Briefly, the sections were deparaffinized in xylene (twice for 10 minutes each), followed by immersion in 100% ethanol (twice for 5 minutes each) before hybridization. Pretreatment was performed according to the Vysis protocol for FFPE tissue specimens. A SOX2-specific DNA probe (green) and a centromere 3-specific probe (red) were used (ZytoLight SPEC SOX2/CEN 3 Dual Color Probe, ZytoVision, Bremerhaven, Germany). The sections were then denatured and hybridized with ThermoBrite (Abbott Molecular, Des Plaines, IL) at 80°C for 5 minutes, followed by an overnight at 37°C. Post-hybridization washes were performed according to the Vysis protocol for FFPE tissue specimens (Abbott Molecular), and DAPI counterstaining was then performed. Semi-quantitative analysis of the SOX2 amplification status was performed by comparing the number of green signals (SOX2 target regions) to the number of red signals in each sample (reference regions). A non-amplified nucleus showed one green target signal for every corresponding red reference signal, with a green/red ratio of 1:1 (Fig. 1A). Cases containing 2-9 more green target signals than red signals in at least 30% of their tumor cells were defined as having low-level SOX2 amplification (Fig. 1B). Cases with an additional ≥ 10 green signals in a cluster-like formation were defined as having high-level SOX2 amplification (Fig. 1C). All tissue slides were analyzed under a 100× oil immersion objective lens using a fluorescence microscope equipped with the appropriate filters. At least 100 nuclei per case were assessed.
3. ImmunohistochemistryFour-micrometer-thick tissue sections were deparaffinized, rehydrated, and washed twice in buffer. The slides were incubated in hydrogen peroxide for 10 minutes to reduce nonspecific background staining due to endogenous peroxidases, and then washed four times in buffer. Primary antibodies against human SOX2 (1:200, R&D Systems, Minneapolis, MN) were then applied, and slides were incubated according to the manufacturer’s recommended protocols. The slides were washed four times in buffer, incubated with primary antibody enhancer for 20 minutes at room temperature, and then washed four times in buffer. Next, horseradish peroxidase polymer was applied to the slides, which were then incubated for 30 minutes at room temperature before washing four times in buffer, followed by incubation with hematoxylin and chromogen, washed four times in deionized water, and counterstained. The staining results were evaluated and each case was scored from 0 to 2, according to the intensity of nuclear staining in the tumor cells. The staining intensity of the normal bronchial epithelium served as the internal control and was given an arbitrary score of 1. Each tumor was compared to the internal control and given a score of 1 (moderate expression) (Fig. 1D) when the intensity was the same as that of the internal control, 2 (strong expression) (Fig. 1E) when stronger, and 0 (weak expression) (Fig. 1F) when weaker.
4. Statistical analysesDisease-free survival (DFS) was estimated from the time of diagnosis to the time of initial tumor relapse (local recurrence or distant) or death from any cause. Overall survival (OS) time was measured from the time of diagnosis to death or last follow-up date. For univariate analysis, Kaplan-Meier survival analyses were used to estimate OS and DFS, and differences in the survival rates were compared using log-rank tests. A Cox proportional hazards model was used for multivariate analysis to evaluate prognostic factors influencing OS and DFS. Multivariate analysis was performed using backwards elimination to stay in the model. Hazard ratios (HR) are given with 95% confidence intervals (95% CI). Correlations between categorical variables were examined using χ2 or Fisher exact tests. Continuous variables were compared to categorical variables using Mann-Whitney U tests. Statistical significance was defined as a p-value of < 0.05 for all analyses. SPSS ver. 20.0 (IBM Co., Armonk, NY) was utilized for all statistical analyses.
Results1. Patient characteristicsThe median patient age was 66 years (range, 48 to 73years), and 32 patients were male (97%). Most patients had an Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1 (93.9%) and a positive smoking history (75.8%). Fourteen patients (42.4%) were classified as T3 or T4, and 27 patients (81.8%) as N2 or N3. A positive margin of resection was found in 11 patients (33.3%), lymphovascular invasion and perineural invasion in seven patients (21.2%) and three patients (9.1%), and extranodal extension in four patients (12.1%). The median total dose and fraction size of radiotherapy were 59.4 Gy (range, 50.4 to 69 Gy) and 1.8 Gy (range, 1.8 to 2 Gy), respectively. The patient characteristics are listed in Table 1.
2. Correlation between SOX2 gene amplification and protein expression and clinicopathological characteristics
SOX2 amplification was assessed using FISH and protein expression was determined using immunohistochemistry in lung squamous cell carcinoma patients (Fig. 1). High- and low-level amplification were observed in four (12.2%) and 18 patients (54.5%), respectively (Table 1). SOX2 gene amplification group was defined as low or high level amplification. Strong and moderate expressions were observed in eight (24.3%) and 18 patients (54.5%), respectively (Table 1). SOX2 overexpression was defined as a score of 1 or 2, and no overexpression was defined as a score of 0.
Analysis of the correlation between SOX2 gene amplification and protein expression showed significant association of SOX2 overexpression with SOX2 gene amplification (p=0.002) (Table 2). Next, we analyzed the association between several clinicopathological characteristics and SOX2 gene amplification and protein expression. However, no clinicopathological factor showed significant association with SOX2 gene amplification or protein expression (Table 3).
3. Prognostic factors for OS and DFSThe median follow-up period for the surviving patients was 58 months (range, 5 to 102 months). The 5-year OS and DFS for all patients was 50.7% and 39.9%, respectively. In prognostic analysis using Kaplan-Meier curves, SOX2 overexpression was the only significant prognostic factor for OS (yes vs. no; median, 73 months vs. 8 months; p=0.01) (Table 4). Kaplan-Meier curves also showed that patients with SOX2 overexpression had significantly better OS, as shown in Fig. 2A. Age and total dose showed better OS with a statistically significant trend. However, SOX2 amplification did not show association with OS (Table 4, Fig. 2B). In multivariate analysis, SOX2 overexpression (HR, 0.1; 95% CI, 0.02 to 0.5; p=0.005) and age (HR, 0.33; 95% CI, 0.11 to 0.98; p=0.046) were the independent prognostic factors for OS (Table 4). In univariate analysis, age (< 65 vs. ≥ 65; median, 96 months vs. 12 months; p=0.02) and total radiation dose (> 59.4 Gy vs. ≤ 59.4 Gy; median, 96 months vs. 30 months; p=0.04) were the significant prognostic factors for DFS (Table 4). SOX2 overexpression showed a significant trend toward better DFS (p=0.08) (Table 4, Fig. 2C). SOX2 amplification also did not show association with DFS (Table 4, Fig. 2D). In multivariate analysis, SOX2 overexpression (HR, 0.15; 95% CI, 0.04 to 0.65; p=0.01) and total radiation dose (HR, 0.13; 95% CI, 0.02 to 0.7; p=0.02) were the independent prognostic factors for DFS (Table 4).
DiscussionSOX2 plays roles in a variety of pathological processes from normal development to cancer. Overexpression of SOX2 is not associated with epithelial cell differentiation in the lungs, but its expression shows a notable increase in basal cell precursors (p63+ Krt14− cells) [10]. Therefore, SOX2 regulates the balance between self-renewal and differentiation by controlling the fate of basal cells and also triggers lung epithelial carcinogenesis [11].
SOX2 play a role in various signal transduction pathways and exerts various biological effects via extensive networks involving upstream and downstream molecules and protein interactions [12]. Many proteins upstream of SOX2 have been identified. EGFR-Src-Akt signaling is relevant to the self-renewal of cancer stem cells and has an effect on side population cells in lung cancer [13]. Several downstream effectors of SOX2 have been identified including those involved in cancer stem cell-related signal transduction pathways. For example, C-MYC, WNT1, WNT2, and NOTCH1 play roles in non-small cell lung cancer cells [14]. SOX2 was linked with apoptosis in non-small cell lung cancer via the MAPK4-survivin pathway [15]. SOX2 forms a network with cell cycle regulators including cyclin D1, cyclin E, and p27, as well as other biologically important pathways such as β-catenin and transforming growth factor β [16-18].
Little is known about the relationship between radioresistance and SOX2. An analysis of the SOX2 interactome using immunoprecipitation coupled with mass spectrometry analysis by Fang et al. [19] found interactions between SOX2 and proteins involved in DNA repair, such as XRCC1, 5, and 6, which were associated with radioresistance in vitro and in human tissues [20-22]. Future studies are needed to investigate the direct relationship between radioresistance and these genes. In addition, cancer stem cell radioresistance can be explained by an increased recovery from DNA damage and a decrease in production of reactive oxygen species [23,24]. Therefore, SOX2 might influence radioresistance through regulation of cancer stem cell activity. If SOX2 increases radioresistance, SOX2 overexpression would result in worse prognosis in irradiated patients, in contrast to our findings. However, all patients enrolled in our study underwent surgery followed by postoperative radiotherapy, and most patients received adjuvant systemic chemotherapy. Thus, we concluded that our findings were not appropriate for investigating the relation between SOX2 and radioresistance. Instead, we suggest that further studies designed for decisive examination of this point, such as a prospective study with lung squamous cell carcinoma patients who receive definitive radiotherapy, are required.
Several previous studies investigated whether SOX2 amplification was associated with prognosis in non-small cell lung cancer. Although some reported that SOX2 amplification showed close correlation with poor prognosis, others reported that patients with amplification had a good survival outcome. These conflicting results arise from the limited availability of patient tissues and methods for investigation of SOX2 amplification. However, our findings showed that SOX2 amplification is not associated with survival.
A meta-analysis utilizing the eight published series found significantly greater expression of SOX2 in squamous carcinoma than in adenocarcinoma, which was associated with improved OS. Similarly, multivariate analysis also showed that SOX2 overexpression is the significant prognostic factor for OS and DFS. Therefore, we suggest that overexpression of SOX2 might be a positive prognostic factor in patients with lung squamous cell carcinoma. However, as several studies showed that the use of small interfering RNA to knockdown SOX2 decreased the growth and radioresistance of cancer cells in contrast to clinical studies [18,25], it remains unclear whether this could result in a feasible therapeutic approach. Therefore, further studies involving well-designed translational research from benchside to bedside are needed.
The current study included a homogenous NSCLC patient population treated with adjuvant radiotherapy. To the best of our knowledge, this is the first study to investigate whether SOX2 overexpression has prognostic significance in patients who underwent radiotherapy in NSCLC, albeit only a small number of patients were included. Nevertheless, despite the high possibility, we cannot assume that postoperative radiotherapy changed the prognosis of patients and that SOX2 is involved in this effect. We were also unable to confirm a relationship between SOX2 and radioresistance. Thus, further studies are certainly needed in order to validate the effect of postoperative radiotherapy on prognosis and the relationship between radiotherapy and SOX2. Still, based on our findings, it is meaningful that the prognostic value of SOX2 overexpression was verified by utilizing the homogenous database of all patients treated with the whole definitive treatment scheme including radical surgery followed by postoperative radiotherapy.
This study has several shortcomings. Primarily, it was retrospective in design and included only a small number of patients who underwent postoperative radiotherapy. We thought that SOX2 overexpression showed a significant trend toward better DFS on the Kaplan-Meier curve in Fig. 2C due to the small number of patients in spite of significant relationship between SOX2 overexpression and progression-free survival in multivariate analysis. A welldesigned large-scale prospective study is necessary to confirm our results. In addition, despite a very strong positive relationship between SOX2 protein overexpression and gene amplification, gene amplification of SOX2 did not show significant association with OS or DFS, in contrast to SOX2 overexpression. Similarly, in a recent large retrospective study, only SOX2 overexpression showed significant association with better overall survival, although significant positive correlation was observed between SOX2 protein overexpression and gene amplification [8]. Furthermore, from an indepth review of our data, five patients with SOX2 overexpression without gene amplification showed an excellent outcome of median OS (60 months). Based on these findings, we suggest that a high SOX2 protein level may affect the prognosis of lung squamous cell carcinoma patients more acutely than SOX2 gene amplification. Consequently, we recommend an evaluation of the mechanism of SOX2 overexpression with no gene amplification. Thus, further studies investigating other mechanisms of SOX2 protein overexpression, such as chromosomal translocation or mutation without gene amplification, would also be necessary.
ConclusionIn conclusion, SOX2 overexpression could be a positive prognostic factor in lung squamous cell carcinoma patients treated with postoperative adjuvant radiotherapy. Further studies are needed to investigate whether SOX2 is associated with radioresistance and how this could be exploited therapeutically.
AcknowledgmentsThis study was supported by a faculty research grant from Yonsei University College of Medicine for 2012(6-2012-0187). We thank Jin-Kyu Park and Korea CFC pathology laboratory to support our study.
Fig. 1.
SOX2 amplification was assessed using fluorescence in situ hybridization (FISH, ×1,000) and protein expression was determined using immunohistochemistry (×200) in lung squamous cell carcinoma patients. SOX2-specific DNA probe in green combined with a centromere 3-specific probe in red was applied for FISH. (A) Nucleus without SOX2 amplification. (B) Nucleus with low-level SOX2 amplification (arrows). (C) Nucleus with high-level SOX2 amplification (arrows). (D) Moderate nuclear SOX2 expression (arrow). (E) Strong nuclear SOX2 expression (arrow). (F) Weak nuclear SOX2 expression. 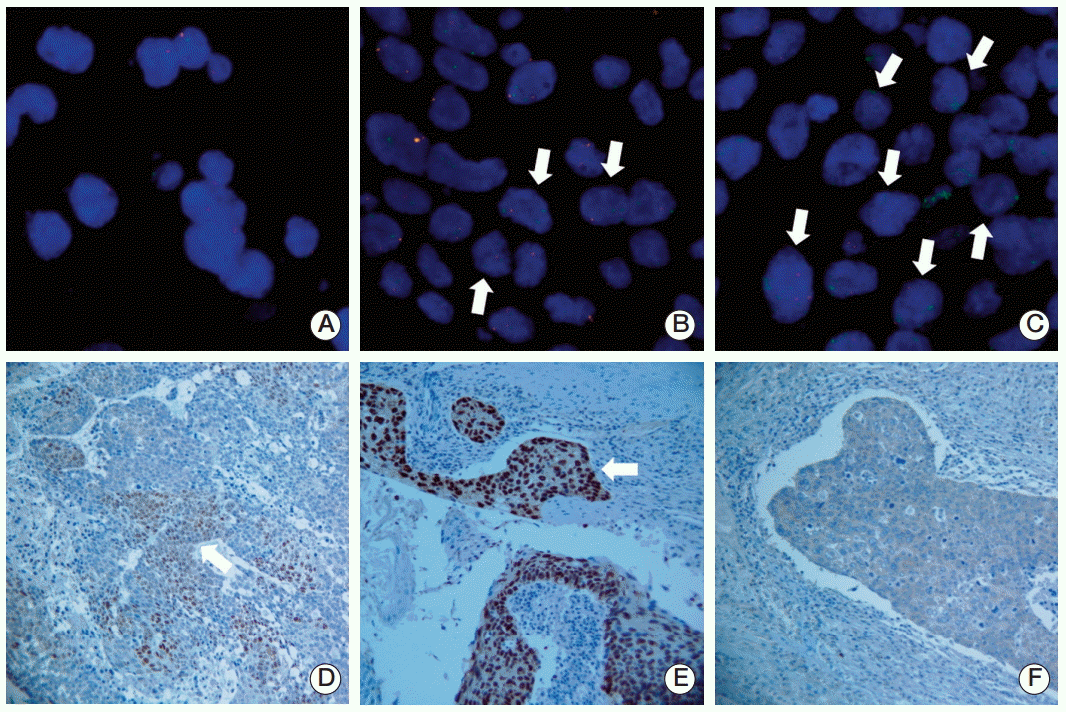
Fig. 2.Kaplan-Meier curves for overall survival and disease-free survival rates. (A) Patients with SOX2 overexpression showed a significantly longer overall survival rate compared to those without (median, 73 months vs. 8 months, respectively; p=0.01). (B) Patients with SOX2 amplification did not show a better overall survival rate than those without amplification(median, 69 months vs. 59 months; p=0.95). (C) Patients with SOX2 overexpression showed a significant trend toward longer disease-free survival compared to those without (median, 57 months vs. 5 months; p=0.08). (D) Patients with SOX2 amplification did not show a longer disease-free survival than those without amplification (median, 30 months vs. 82 months; p=0.48). 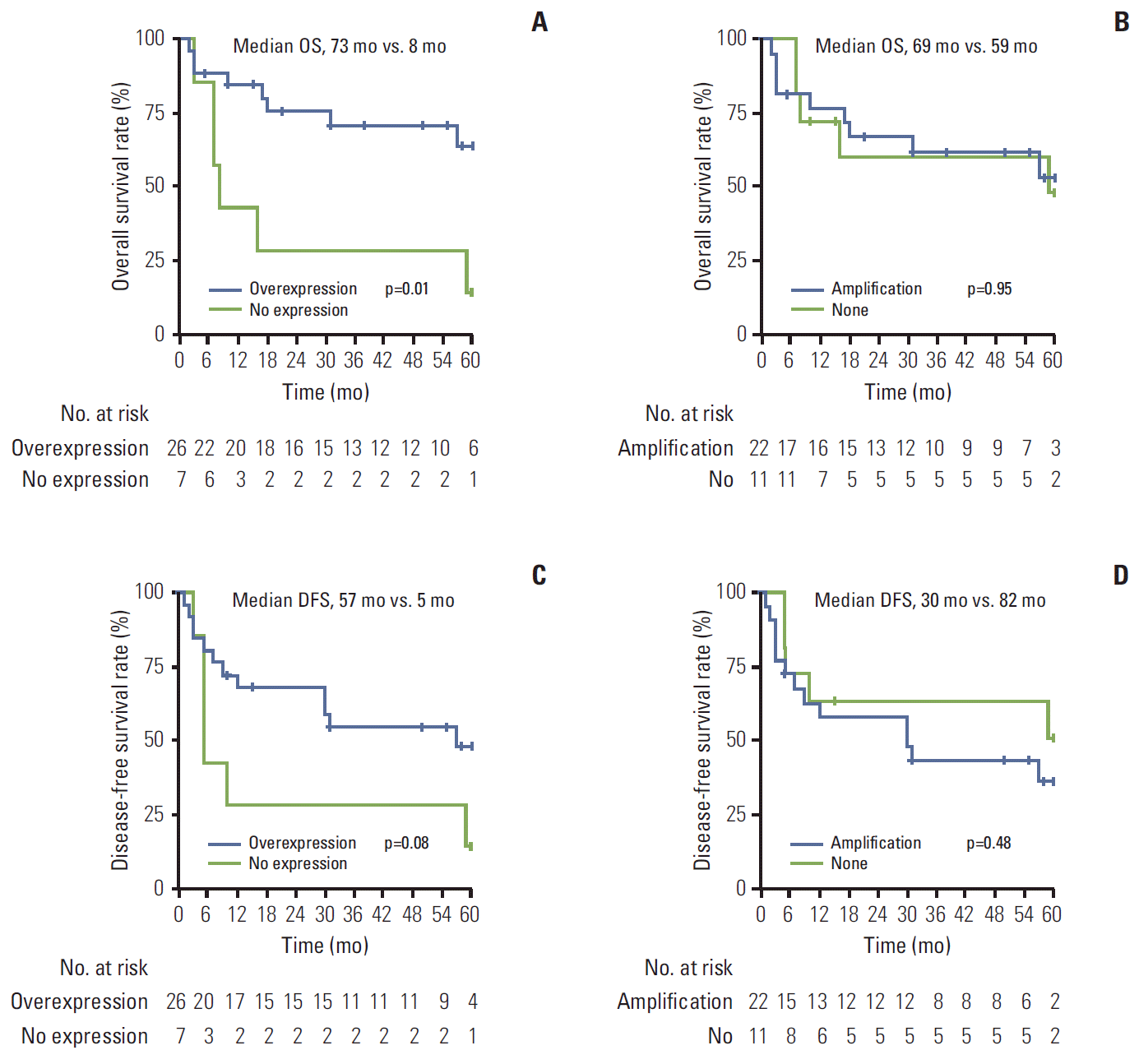
Table 1.Patient characteristics Table 2.Relationship between SOX2 gene amplification and SOX2 protein expression
Table 3.Clinicopathologic characteristics according to SOX2 amplification and expression Table 4.Stepwise uni- and multi-variate analysis using Cox regression model for overall survival and disease-free survival References1. Jung KW, Park S, Won YJ, Kong HJ, Lee JY, Seo HG, et al. Prediction of cancer incidence and mortality in Korea, 2012. Cancer Res Treat. 2012;44:25–31.
2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
3. Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–42.
5. Long KB, Hornick JL. SOX2 is highly expressed in squamous cell carcinomas of the gastrointestinal tract. Hum Pathol. 2009;40:1768–73.
6. Yuan P, Kadara H, Behrens C, Tang X, Woods D, Solis LM, et al. Sex determining region Y-Box 2 (SOX2) is a potential cell-lineage gene highly expressed in the pathogenesis of squamous cell carcinomas of the lung. PLoS One. 2010;5:e9112
7. Velcheti V, Schalper K, Yao X, Cheng H, Kocoglu M, Dhodapkar K, et al. High SOX2 levels predict better outcome in non-small cell lung carcinomas. PLoS One. 2013;8:e61427
8. Wilbertz T, Wagner P, Petersen K, Stiedl AC, Scheble VJ, Maier S, et al. SOX2 gene amplification and protein overexpression are associated with better outcome in squamous cell lung cancer. Mod Pathol. 2011;24:944–53.
9. Greene FL, Page DL, Fleming ID, Fritz AG, Balch CM, Haller DG, et al. AJCC cancer staging manual. 6th edNew York: Springer; 2002.
10. Gontan C, de Munck A, Vermeij M, Grosveld F, Tibboel D, Rottier R. Sox2 is important for two crucial processes in lung development: branching morphogenesis and epithelial cell differentiation. Dev Biol. 2008;317:296–309.
11. Hussenet T, du Manoir S. SOX2 in squamous cell carcinoma: amplifying a pleiotropic oncogene along carcinogenesis. Cell Cycle. 2010;9:1480–6.
12. Liu K, Lin B, Zhao M, Yang X, Chen M, Gao A, et al. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal. 2013;25:1264–71.
13. Singh S, Trevino J, Bora-Singhal N, Coppola D, Haura E, Altiok S, et al. EGFR/Src/Akt signaling modulates Sox2 expression and self-renewal of stem-like side-population cells in non-small cell lung cancer. Mol Cancer. 2012;11:73.
14. Chen S, Xu Y, Chen Y, Li X, Mou W, Wang L, et al. SOX2 gene regulates the transcriptional network of oncogenes and affects tumorigenesis of human lung cancer cells. PLoS One. 2012;7:e36326
15. Chen S, Li X, Lu D, Xu Y, Mou W, Wang L, et al. SOX2 regulates apoptosis through MAP4K4-survivin signaling pathway in human lung cancer cells. Carcinogenesis. 2014;35:613–23.
16. Wu F, Zhang J, Wang P, Ye X, Jung K, Bone KM, et al. Identification of two novel phenotypically distinct breast cancer cell subsets based on Sox2 transcription activity. Cell Signal. 2012;24:1989–98.
17. Lin F, Lin P, Zhao D, Chen Y, Xiao L, Qin W, et al. Sox2 targets cyclinE, p27 and survivin to regulate androgen-independent human prostate cancer cell proliferation and apoptosis. Cell Prolif. 2012;45:207–16.
18. Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, et al. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem. 2008;283:17969–78.
19. Fang X, Yoon JG, Li L, Tsai YS, Zheng S, Hood L, et al. Landscape of the SOX2 protein-protein interactome. Proteomics. 2011;11:921–34.
20. Niu Y, Zhang X, Zheng Y, Zhang R. XRCC1 deficiency increased the DNA damage induced by gamma-ray in HepG2 cell: involvement of DSB repair and cell cycle arrest. Environ Toxicol Pharmacol. 2013;36:311–9.
21. Chang HW, Kim SY, Yi SL, Son SH, Song DY, Moon SY. Expression of Ku80 correlates with sensitivities to radiation in cancer cell lines of the head and neck. Oral Oncol. 2006;42:979–86.
22. Ayene IS, Ford LP, Koch CJ. Ku protein targeting by Ku70 small interfering RNA enhances human cancer cell response to topoisomerase II inhibitor and gamma radiation. Mol Cancer Ther. 2005;4:529–36.
23. Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||