AbstractPurposeThe peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor that regulates expression of mediators of lipid metabolism and the inflammatory response. Thyroid hormone receptor-associated proteins 220 (TRAP220) is an essential component of the TRAP/Mediator complex. The objective of this study was to clarify whether PPARγ or TRAP220 are significant prognostic markers in resectable colorectal cancer (CRC).
Materials and MethodsA total of 399 patients who underwent curative resection for CRC were enrolled. We investigated the presence of PPARγ and TARP220 in CRC tissues and adjacent normal tissues by immunohistochemistry. Correlation between the expression of these factors and clinicopathologic features and survival was investigated.
ResultsMedian age of the patients was 63 years (range, 22 to 87 years), and median follow-up duration 61.1 months (range, 2 to 114 months). PPARγ and TRAP220 expression showed significant correlation with depth of invasion (p=0.013 and p=0.001, respectively). Expression of TRAP220 also showed association with lymph node metastasis and TNM stage (p=0.001). Compared with patients with TRAP220 negative tumors, patients with TRAP220 positive tumors had longer 5-year disease-free survival (DFS) tendency (p=0.051). Patients who were PPARγ positive combined with TRAP220 positive had a better 5-year DFS (64.8% vs. 79.3%, p=0.013). In multivariate analysis expression of both PPARγ and TRAP220 significantly affected DFS (hazard ratio, 0.620; 95% confidence interval, 0.379 to 0.997; p=0.048).
IntroductionColorectal cancer (CRC) is the third most commonly diagnosed cancer in males and the second in females, with over 1.2 million new cancer cases and 608,700 estimated deaths in 2008 [1], and it is the fourth most frequent cause of cancer-specific death in Korea [2]. Assessment of biological prognostic factors is of clinical importance in CRC. The outcome of cancer patients may be influenced by variability in tumor biology. Thus, tumors with similar clinical or pathologic characteristics frequently show a different clinical outcome. Insight into the underlying molecular mechanisms of initiation and progression of CRC is needed in order to identify groups of patients with a poor prognosis.
Despite the usefulness of American Joint Committee on Cancer (AJCC) TNM classification for staging of CRC patients and selection for specific treatment, it is not a completely sufficient method, because many patients at the same stage may have various clinical outcomes, thus rendering the conventional staging system incapable of precisely predicting prognoses. Therefore, there is a great need to identify the molecular markers of more aggressive CRC for selection of patients for adjuvant systemic or targeted therapies. In this regard, many studies have focused on biomarkers that perform a critical role in CRC [3]. For these factors to be clinically useful, they should be routinely available, well standardized and validated in different patient cohorts. However, only a few molecular-based factors have been used in routine clinical practice. There remains a continuing need to identify clinically relevant factors that would improve the prediction of survival in patients undergoing potentially curative surgery for CRC.
TRAP/Mediator, an evolutionarily conserved multisubunit coactivator complex [4], acts as a molecular bridge between gene-specific transactivators and the RNA polymerase II associated basal transcription machinery. TRAP/Mediator consists of 25-30 subunits that are thought to assemble in relatively discrete modules. Thyroid hormone receptor-associated proteins 220 (TRAP220), the main subunit of the TRAP/Mediator complex, binds to nuclear hormone receptors in the presence of their cognate ligand, as a cofactor of transcription machinery [5]. TRAP/Mediator may be involved in a variety of human cancers [6,7]. TRAP220 plays an important role in the development and progression of lung adenocarcinoma [6]. TRAP220 promoter methylation and gene silencing occur in sporadic CRC patients, and the finding that TRAP220 was hypermethylated in CRC patients in both tumors and matched normal mucosa implies that hypermethylation of this gene occurs early in colorectal carcinogenesis and appears to be associated with tumor initiation rather than tumor progression [7].
Peroxisome proliferator-activated receptors (PPARs) are members of the nuclear hormone receptor superfamily. PPARs, which are ligand-activated transcription factors, have three different isoforms: PPARα, PPARβ/δ, and PPARγ [8]. PPARγ constitutes the most extensively studied of the three PPAR-subtypes to date, since it plays a crucial role in glucose and lipid homeostasis, as well as cell differentiation, apoptosis, and cancer [9].
However, conflicting results have been reported so far, raising the question as to whether PPARγ facilitates or suppresses tumorigenesis [10]. Several studies which reported PPARγ expression in CRC [11-14], have shown that sporadic CRCs presenting reduced PPARγ expression levels are significantly associated with patients' worse prognosis [11]. One recent large cohort study suggested that PPARγ expression is independently associated with good prognosis in CRC [12]. Other studies did not show a prognostic value of tumoral PPARγ status, but were limited sizes [13,14]. Thus, clinical significance of PPARγ expression in human CRC remains uncertain.
Significant association was observed between expression of TRAP220 and expression of PPARγ. The nuclear receptor-interacting TRAP220 subunit of Mediator is required for PPARγ stimulated adipogenesis of mouse embryonic fibroblasts [15]. TRAP220 modestly increased the transcriptional activity with a PPARγ responsive reporter, and a fragment of TRAP220 spanning the two LXXLL motifs acted as a dominant-negative repressor, suggesting that TRAP220 is a coactivator of PPARγ [16].
In the current study, we examined the expression of TRAP220 and PPARγ in 399 cases of human colon cancer using immunohistochemical technique and correlated it with clinicopathological parameters. We wanted to determine whether the immunohistochemical expression of TRAP220 and PPARγ could provide useful information as a novel prognostic option for treatment of CRC.
Materials and Methods1. PatientsA total of 399 patients were included in this study between May 2005 and December 2008. All patients had histologically confirmed adenocarcinomas of the colon or rectum, and had undergone potentially curative resections, with neither gross nor microscopic evidence of residual disease. Patients with familial adenomatous polyposis or inflammatory bowel disease, synchronous colorectal or extracolorectal cancers, and those lost to follow-up were excluded. None of the patients had a family history of CRC or received preoperative chemotherapy or radiotherapy. Staging was based on routine postoperative histopathological analysis and clinical assessment by the AJCC TNM staging system, sixth edition. Tissue samples from patients were formalin-fixed and paraffin-embedded.
Patients were followed up every 3 months for the first 2 years, every 6 months for the next 3 years, and annually thereafter. Patients underwent physical examination and serum carcinoembryonic antigen test at each follow-up; underwent full colonoscopy the first year from surgery, then once every 3 to 5 years if no polyp was identified. Abdominal computed tomography (CT) scans were obtained generally every 6-12 months. Chest plain film or CT scan was performed generally every 6-12 months. Clinical outcomes were followed from the date of surgery to either the date of death or January 2012. Hospital records were used to accurately identify the length of survival. The study was approved by the Institutional Review Board (DAUH-IRB-12-168).
2. Construction of tissue microarrayOne-millimeter cores were removed from the CRC samples that had previously been formalin-fixed and paraffin-embedded. For all of the arrays, three cores of different areas of the tumor were removed from each case and placed in a new blank recipient paraffin block in a previously described manner [17], and 4-μm-thick sections were taken for all of the immunohistochemical staining. Full cross-sections from the paraffin blocks were used for five of the CRCs, along with the adjacent normal colorectal tissue, to confirm the staining patterns observed on the tissue microarrays.
3. ImmunohistochemistryImmunohistochemical staining for PPARγ and TRAP220 was performed on the tissue microarray slides using the avidin-biotin-peroxidase complex method. All sections were deparaffinized through a series of xylene baths, and rehydration was performed with a series of graded alcohol solutions. To enhance the immunoreactivity, microwave antigen retrieval was performed at 750 W for 30 minutes in Tris EDTA (pH 9.0). After blocking the endogenous peroxidase activity with 5% hydrogen peroxidase for 10 minutes, incubation with the primary antibody was performed for 1 hour at room temperature. The primary antibodies used in immunostaining were rabbit polyclonal antibody directed against TRAP220 (CRSP1, Novus Biologicals, Littleton, CO) used at a 1:250 dilution and mouse monoclonal antibody directed against PPARγ (Santa Cruz Biotechnology, Santa Cruz, CA) used at a 1:50 dilution. An EnvisionChem Detection Kit (DakoCytomation, Carpinteria, CA) was used for the secondary antibody at room temperature for 30 minutes. After washing the tissue samples in Tris buffered saline for 10 minutes, 3,3′-diaminobenzidine was used as a chromogen, followed by application of Mayer’s hematoxylin as a counterstain. Positive controls for TRAP220 and PPARγwere normal adrenal gland and normal breast tissue, respectively. Negative control was obtained by substituting primary antibody with buffer.
4. Immnohistochemical assessmentThe percentage and intensity of the immunoreactive tumor cells in each core were recorded and the final value of the positive tumor cells was determined as the mean of the immunoreactivity of the three cores. The presence of tumor tissue in at least two interpretable cores was required for inclusion of a case in the statistical analysis. All slides were independently evaluated by an experienced pathologist (M.S.R.) with no knowledge of any of the clinicopathologic data. There were only minor discrepancies in the evaluation. Slides with discrepant evaluation were reevaluated under a multi-head microscope until a consensus evaluation was obtained.
PPARγ immunoreactivity was defined as those showing a nuclear staining pattern of the tumor tissue. TRAP220 immunoreactivity was defined as those showing a nuclear with/without cytoplasmic staining pattern of the tumor tissue with minimal staining background. The percentage scoring of the immunoreactive tumor cells was as follows: 0 (0%), 1 (1%-10%), 2 (11%-50%), and 3 (> 50%). The staining intensity was visually scored and stratified as follows: 0 (negative), 1 (weak, if it was a blush), and 2 (strong, if it was obviously positive at 20× magnification). A final score was obtained for each case by multiplying the percentage and the intensity score. Therefore, tumors with multiplied score exceeding 4 (i.e., tumors with a strong intensity of > 10% of the tumor cells) were recorded as having positive immunoreactivity for PPARγ (Fig. 1) and TRAP220 (Fig. 2); all other scores were considered negative.
5. Statistical analysisThe associations between PPARγ or TRAP220 and the clinicopathologic parameters (sex, age, tumor size, grade, depth of bowel wall invasion, lymph node metastasis, TNM stage, lymphovascular invasion) were assessed via chi-square or Fisher exact tests. Disease-free survival (DFS) was defined as the length of time from surgery to initial disease recurrence. Overall survival was defined as the length of time from surgery to death. The Kaplan-Meier method was utilized in construction of curves for DFS and overall survival (OS). Data on patients who died without evidence of disease recurrence were censored at the time of death for DFS calculations. The log-rank test was employed to compare distributions. Cox’s proportional hazard analysis via a stepwise procedure was used to determine the independent factors that were significantly related to patient prognosis. All tests were two-sided, and p-values of < 0.05 were considered statistically significant. Analyses were performed using IBM SPSS Statistics ver. 19.0 (IBM Corp., Armonk, NY).
Results1. Patient characteristicsPatient’s characteristics are shown in Table 1. Of the 399 CRC patients, 223 (55.9%) were men, and the median patient age was 63 years (range, 22 to 87 years); 200 patients (50.1%) had colon cancer, and 151 (37.8%) had rectal cancer. All patients had adenocarcinomas, most of them were well (6.9%) and moderately (32.6%) differentiated cancer. Only 58 patients (13.6%) had T1 or T2 lesions, and 219 patients (54.9%) were lymph node negative. The postoperative stages were II, III, and IV in 217 (544%), 168 (42.1%), and 14 (35%) patients, respectively. All patients underwent surgical resection, and 70 (17.5%) had lymphovascular invasion.
2. PPARγ and TRAP220 expressionPositive PPARγ expression was found in 166 of 399 cases (41.6%) of CRC examined, while it was negative in the remaining 233 cases (58.4%). TRAP220 immunoreactivity was observed in 170 cases (42.6%). Similar staining characteristics were observed for all cores for each tumor. The staining patterns of the tissue microarray cores showed concordant results with those of the five full cross-sections. The pattern of PPARγ positive staining was detected in the nuclei. Expression of TRAP220 protein was detected in the nuclei with/without cytoplasm of both normal cells and tumor cells. The frequency of PPARγ expression in cases with TRAP220 expression was significantly higher than that in cases without TRAP220 expression (p < 0.001).
3. Correlations between expression of PPARγ and TRAP 220 and clinicopathologic parametersPPARγ expression showed significant correlation with depth of invasion (p=0.013). Expression of PPARγ protein was not affected either by age in male and female patients, or by tumor location. No statistically significant difference in PPARγ expression was found among cases with different TNM stage. The various clinicopathologic characteristics of the patients and their tumors were compared according to TRAP220 immunoreactivity. TRAP220 also showed significant correlation with depth of invasion (p=0.011). Tumors with a negative TRAP220 expression more frequently showed lymph node metastasis (p=0.001) and an advanced TNM stage (p=0.011). No significant association was found between TRAP220 expression and the other clinicopathologic characteristics.
4. Expression of PPARγ and TRAP220 and clinical outcomesThe median follow-up duration was 61.1 months (range, 2 to 114 months). The relationship between clinicopathological characteristics and survival in patients undergoing potentially curative resection for CRC is shown in Table 2. In univariate analysis, site of primary tumor (p < 0.001), lymphovascular invasion (p < 0.001), depth of invasion (p=0.002), lymph node metastasis (p < 0.001), and TNM stage (p < 0.001) showed significant association with 5-year DFS. Patients with negative TRAP220 cancers showed lower DFS than patients with positive TRAP220 (64.2% vs. 73.6%, p=0.051) (Fig. 3A). However, expression of PPARγ was not associated with DFS (65.5% vs. 72.4%, p=0.213) (Fig. 4A). Patients who were PPARγ positive combined with TRAP220 positive had a better 5-year DFS (64.8% vs. 79.3%, p=0.013) (Fig. 5A).
Lymphovascular invasion (p < 0.001), depth of invasion (p < 0.001), lymph node metastasis (p < 0.001), and TNM stage (p < 0.001) also showed significant association with 5-year OS. On the other hand, PPARγ and TRAP220 expression were not correlated with 5-year OS (p=0.792 [Fig. 3B], p=0.203 [Fig. 4B], respectively). Patients with PPARγ positive combined with TRAP220 positive expressing cancers showed longer 5-year OS rates compared to those without (71.6% vs. 80.2%, p=0.059) (Fig. 5B). The other clinicopathologic factors were not significant.
The impact of each clinicopathological feature on patients’ survival was assessed using Cox regression analysis (Table 3). Accordingly, TNM stage (p < 0.001) and lymphovascular invasion (p=0.035) were independent prognostic factors of DFS. PPARγ and TRAP220 expression was an independent good prognostic factor (hazard ratio, 0.602; 95% confidence interval, 0.368 to 0.985; p=0.043). TNM stage (p < 0.001) and lymphovascular invasion (p=0.011) were identified as significant predictors of OS, after controlling for the other clinicopathologic parameters. In univariate and multivariate survival analysis, PPARγ and TRAP220 co-expression was a significant independent prognostic factor of 5-year DFS.
DiscussionCRC is one of the most common cancers worldwide. Recent advances in the understanding of CRC have raised expectations that this growing knowledge might lead to cancer prevention. In addition, epidemiologic evidence has demonstrated a link between dietary fat and obesity and an increased risk of CRC [18]. And it was shown that lipid derived molecules could activate the nuclear receptor—PPARγ and induce differentiation [19]. Concerns about identification of genetic alterations that underlie the initiation of CRCs and the link between dietary fat and CRC prompted a significant interest in a potential role of PPARγ in CRC. Several studies have reported on PPARγ expression in CRC [11-14], but the clinical significance of PPARγ expression remains uncertain.
Two previous studies investigated the role of PPARγ expression in patient survival [13,14]. However owing perhaps in part to the small sample size, neither of these studies found an association between PPARγ expression and patient outcome. However, Ogino et al. [12], using 2 large, prospective, cohort-based studies, reported that the expression of PPARγ in tumors is associated with increased survival compared with PPARγ-negative tumors. In the current study, we compared PPARγ expression and clinical features and patient outcomes. In examination of 399 patients with CRC, positive expression of PPARγ was noted in 41.6%. No significant correlation was found between PPARγ expression and age, sex, tumor location, histopathological grade, lymph node metastasis, or TNM stage (Table 1). However PPARγ showed significant correlation with depth of invasion (p=0.013). Patients with tumors negative for PPARγ expression had a somewhat decreased 5-year DFS rate (65.5%) compared to those (73.6%) whose tumors showed positive staining (p=0.213). However PPARγ expression was not correlated with 5-year OS (p=0.792) (Fig. 3A). Further investigation will be required to determine the role of PPARγ in CRC, but Ogino et al. [12] suggested that PPARγ expression is independently associated with good prognosis in CRC.
PPARγ and other nuclear hormone receptors comprise a superfamily of DNA binding transcription factors and also require various transcriptional coactivators for activation, in a ligand-dependent manner, transcription of the specific target genes important for cell growth homeostasis, and differentiation [20]. TRAP220 is a coactivator for PPARγ [16]. However the role of TRAP220 is still questionable. To the best of our knowledge, this is the first study to examine the relationship between TRAP220 expression and the clinicopathological features of CRC. In this study, TRAP220 showed significant correlation with depth of invasion (p=0.011). Tumors with negative TRAP220 expression more frequently showed lymph node metastasis (p=0.001) and an advanced TNM stage (p=0.011). TNM stage and nodal involvement are recognized as prognostic indicators in CRCs, thus TRAP220 may be a valuable marker for nodal metastasis and TNM stage. Although PPARγ and TRAP220 expression did not show correlation with 5-year OS, respectively, PPARγ positive combined with TRAP220 positive expression showed longer 5-year DFS as compared to those without (79.3% vs. 64.8%, p=0.013). The results of univariate and multivariate survival analysis showed that PPARγ and TRAP220 expression was a significant independent prognostic factor of 5-year DFS.
The molecular mechanisms for the antitumor effect of PPARγ activation remain incompletely elucidated. DNA microarray studies show that PPARγ ligand treatment is associated with change of gene expression involved in apoptosis, cell proliferation, and angiogenesis in colon cancer cells [21,22]. Kim et al. [23] found that the loss of TRAP220 expression was associated with increased rates of invasion and metastasis in non-small-cell lung cancer patients. And, consistent with lung cancer patient data, they found that the loss of TRAP220 increases the invasive potential of non-small-cell lung cancer cells by modulating the expression of metastasis-related genes. On the basis of the results, they suggested that TRAP220 may act as a suppressor of human lung cancer metastasis and suggested the need for future studies on the role of TRAP220 in other types of human cancers.
ConclusionWe have demonstrated PPARγ and TRAP220 expression in CRCs using immunohistochemistry. Positive PPARγ and TRAP220 expression was detected in 41.6% and 42.6%, respectively. Although PPARγ and TRAP220 expression did not show correlation with any clinicopathological parameters evaluated, patients who were PPARγ positive combined with TRAP220 positive had a better 5-year DFS (64.8% vs. 79.3%, p=0.013). Thus co-expression of PPARγ and TRAP220 represents a biomarker for good prognosis in CRC patients.
Fig. 1.Immunohistochemical staining in peroxisome proliferator-activated receptor γ (PPARγ) colorectal adenocarcinoma. Expression of PPARγ protein was decided as negative (A) and positive (B). 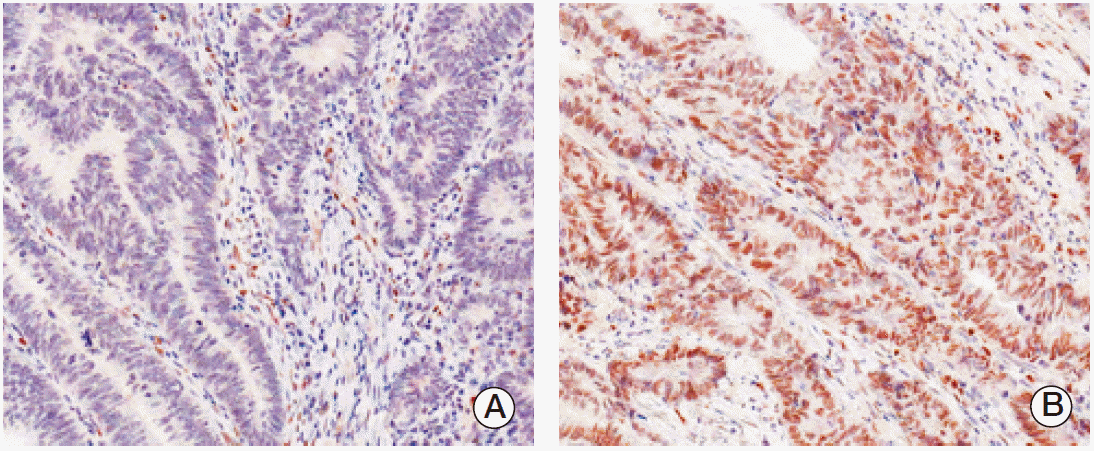
Fig. 2.Immunohistochemical staining of thyroid hormone receptor-associated proteins 220 (TRAP220) in colorectal adenocarcinoma. Expression of TRAP220 protein was decided as negative (A) and positive (B). 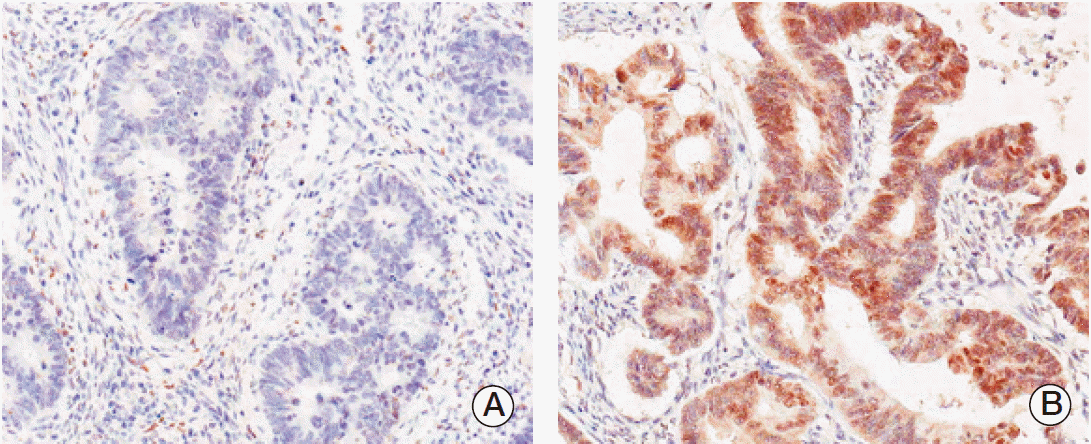
Fig. 3.Kaplan-Meier disease-free survival (A) and overall survival (B) curves in colorectal cancer according to peroxisome proliferator-activated receptor γ (PPARγ) expression. 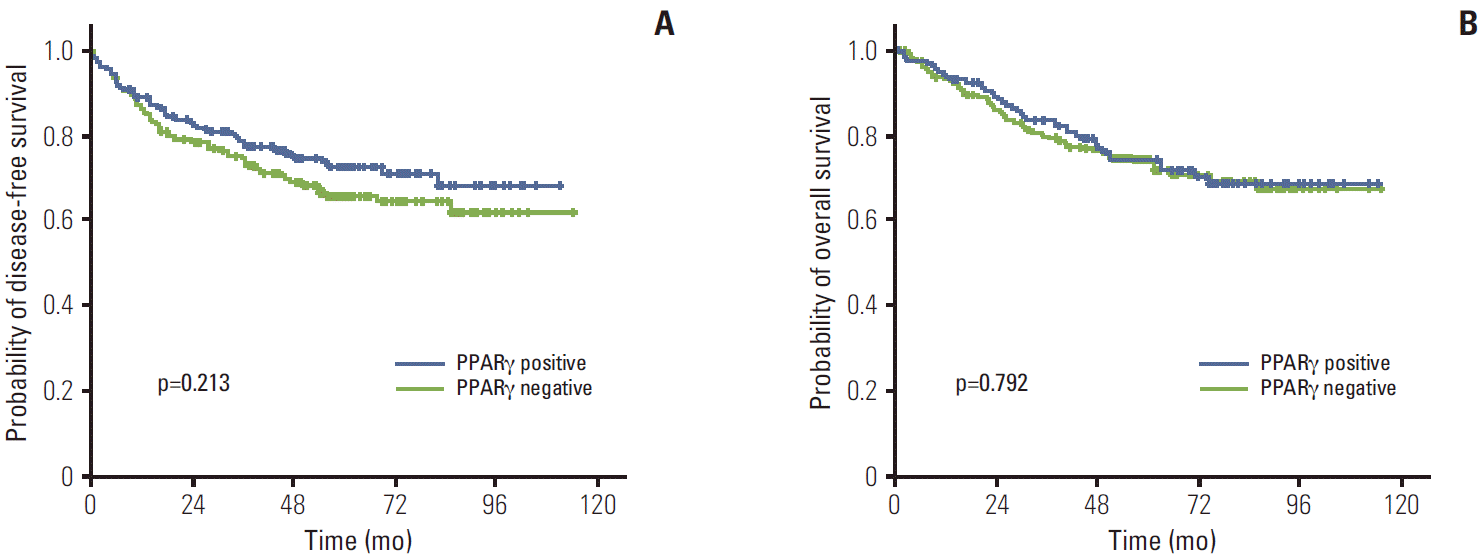
Fig. 4.Kaplan-Meier disease-free survival (A) and overall survival (B) curves in colorectal cancer according to thyroid hormone receptor-associated proteins 220 (TRAP220) expression. 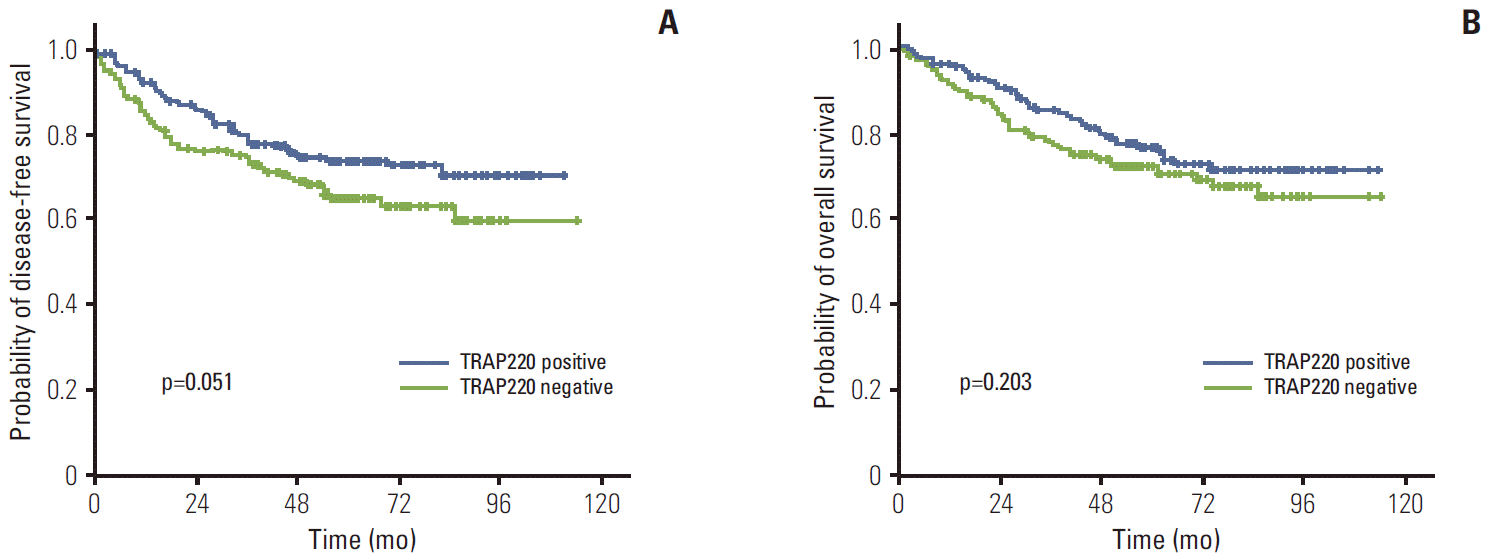
Fig. 5.Kaplan-Meier disease-free survival (A) and overall survival (B) curves in colorectal cancer according to peroxisome proliferator-activated receptor γ and thyroid hormone receptor-associated proteins 220 co-expression. 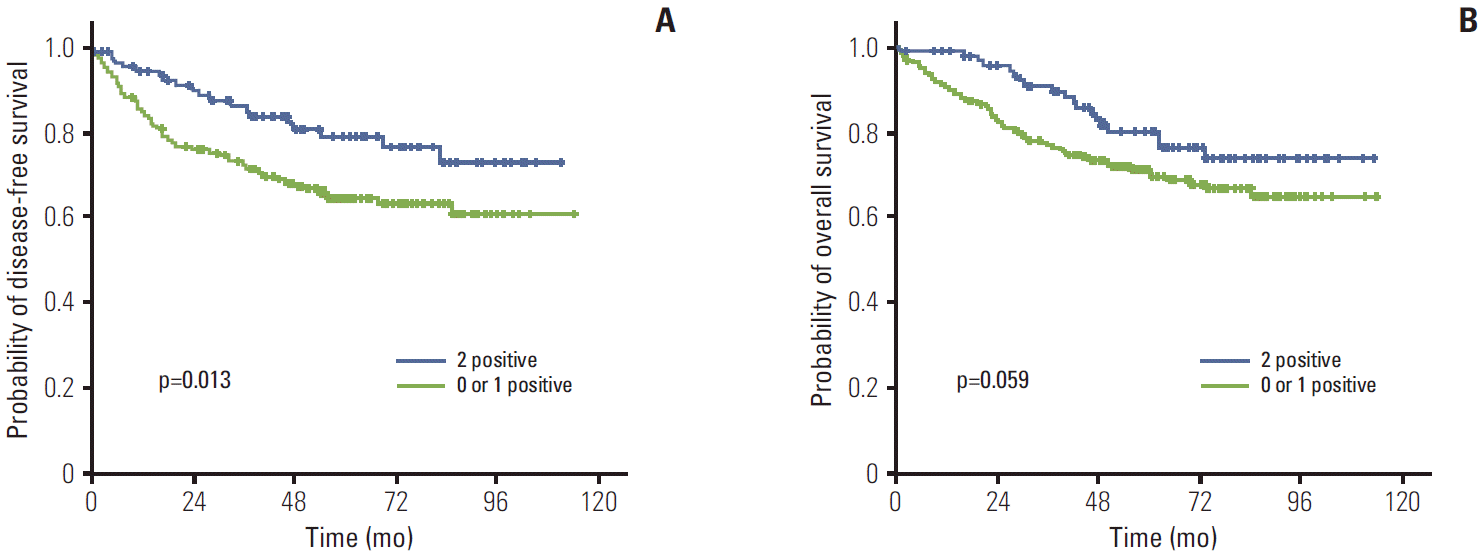
Table 1.Correlation of expression of PPARγ, TRAP220 and clinicopathological findings Table 2.Univariate analysis according to the clinicopathologic findings Table 3.Multivariate analysis of survival References1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
2. Jung KW, Park S, Kong HJ, Won YJ, Lee JY, Seo HG, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2009. Cancer Res Treat. 2012;44:11–24.
3. Pritchard CC, Grady WM. Colorectal cancer molecular biology moves into clinical practice. Gut. 2011;60:116–29.
4. Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–63.
5. Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci U S A. 1998;95:7939–44.
6. Yun J, Son CH, Um SJ, Kwon HC, Lee KE, Choi PJ, et al. A different TRAP220 expression in distinct histologic subtypes of lung adenocarcinoma and the prognostic significance. Lung Cancer. 2011;71:312–8.
7. Howard JH, Frolov A, Tzeng CW, Stewart A, Midzak A, Majmundar A, et al. Epigenetic downregulation of the DNA repair gene MED1/MBD4 in colorectal and ovarian cancer. Cancer Biol Ther. 2009;8:94–100.
8. Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–43.
9. Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. J Med Chem. 2000;43:527–50.
10. Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers: complex stories. Nat Rev Cancer. 2004;4:61–70.
11. Pancione M, Forte N, Sabatino L, Tomaselli E, Parente D, Febbraro A, et al. Reduced beta-catenin and peroxisome proliferator-activated receptor-gamma expression levels are associated with colorectal cancer metastatic progression: correlation with tumor-associated macrophages, cyclooxygenase 2, and patient outcome. Hum Pathol. 2009;40:714–25.
12. Ogino S, Shima K, Baba Y, Nosho K, Irahara N, Kure S, et al. Colorectal cancer expression of peroxisome proliferator-activated receptor gamma (PPARG, PPARgamma) is associated with good prognosis. Gastroenterology. 2009;136:1242–50.
13. Theocharis S, Giaginis C, Parasi A, Margeli A, Kakisis J, Agapitos E, et al. Expression of peroxisome proliferator-activated receptor-gamma in colon cancer: correlation with histopathological parameters, cell cycle-related molecules, and patients' survival. Dig Dis Sci. 2007;52:2305–11.
14. Gustafsson A, Hansson E, Kressner U, Nordgren S, Andersson M, Wang W, et al. EP1-4 subtype, COX and PPAR gamma receptor expression in colorectal cancer in prediction of disease-specific mortality. Int J Cancer. 2007;121:232–40.
15. Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, et al. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–7.
16. Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272:25500–6.
17. Hsu FD, Nielsen TO, Alkushi A, Dupuis B, Huntsman D, Liu CL, et al. Tissue microarrays are an effective quality assurance tool for diagnostic immunohistochemistry. Mod Pathol. 2002;15:1374–80.
18. Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348:1625–38.
19. Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc Natl Acad Sci U S A. 1997;94:4318–23.
20. Rosenfeld MG, Glass CK, et al. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem. 2001;276:36865–8.
21. Qiao L, Li GH, Dai Y, Wang J, Li Z, Zou B, et al. Gene expression profile in colon cancer cells with respect to XIAP expression status. Int J Colorectal Dis. 2009;24:245–60.
|
|
|||||||||||||||||||||||||||||||||||||||||