AbstractPurposeFibroblast growth factor receptor 4 (FGFR4) plays an important role in cancer progression during tumor proliferation, invasion, and metastasis. This study evaluated the prognostic role of FGFR4 polymorphism in patients with resected colon cancer, including the underlying mechanism.
Materials and Methods
FGFR4 polymorphism was characterized in patientswho received curative resection for stage III colon cancer. FGFR4-dependent signal pathways involving cell proliferation, invasion, and migration according to genotypes were also evaluated in transfected colon cancer cell lines.
ResultsAmong a total of 273 patients, the GG of FGFR4 showed significantly better overall survival than the AG or AA, regardless of adjuvant treatment. In the group of AG or AA, combination of folinic acid, fluorouracil, and oxaliplatin (FOLFOX) resulted in better survival than fluorouracil/leucovorin or no adjuvant chemotherapy. However, in GG, there was no difference among treatment regimens. Using multivariate analyses, the Arg388 carriers, together with age, N stage, poor differentiation, absence of a lymphocyte response, and no adjuvant chemotherapy, had a significantly worse OS than patients with the Gly388 allele. In transfected colon cancer cells, overexpression of Arg388 significantly increased cell proliferation and changes in epithelial to mesenchymal transition markers compared with cells overexpressing the Gly388 allele.
IntroductionSurgery is the main treatment for colon cancer, although the recurrence rate is still high. The efficacy of adjuvant chemotherapy treatments has remained constant since introduction of a combination of oxaliplatin and fluorouracil (5-FU) to treat stage III colon cancer. Aside from pathological findings, the lack of biomarkers has made it difficult to identify high risk patients. Furthermore, palliative chemotherapy involving anti–epidermal growth factor or anti–vascular endothelial growth factor has failed to show a significant benefit in clinical trials. Therefore, it is essential to better characterize the molecular mechanisms of colon cancer to develop more effective treatment.
The tumor microenvironment provides the necessary signals for growth and survival of the primary tumor and enhance its invasion and dissemination to distant organs. Targeting tumor cells and the tumor microenvironment is thus crucial to the control and eradication of cancer. The results of extensive studies have suggested that kinase inhibitors to multiple tyrosine residues that target rate-limiting steps in the metabolic pathways of tumor cells may be an effective treatment. Fibroblast growth factors constitute one class of possible targeting agents. These factors bind to four receptors (FGFR1-4) with tyrosine kinase activity involved in epithelial cell growth, migration/metastasis, and angiogenesis [1,2]. Among these receptors, FGFR4 has recently received a great deal of attention [1,3,4]. The overexpression of FGFR4 has been associated with cancer metastasis and poor survival outcome in gastric cancer, lung cancer, breast adenocarcinoma, and rhabdomyosarcoma [5-7]. The role of FGFR4 in colon cancer has been associated with enhancement of tumor cell proliferation, induction of the epithelial-mesenchymal transition (EMT) and resistance to chemotherapy [8-10]. A common polymorphism of FGFR4 involving conversion of guanine to adenine at position 1217 in exon 9 results in the substitution of arginine for glycine at codon 388 (Arg388) in the transmembrane domain, and this polymorphism has several clinical impacts on survival in breast cancer, high grade soft tissue sarcoma, head and neck cancer, and lung and colorectal cancer [10-13].
Thussbas et al. [6] reported poor disease-free survival (DFS) for breast cancer patients with the Arg388 allele of FGFR4 compared to patients with the Gly388 allele of FGFR4 who were treated with surgery followed by adjuvant chemotherapy without a difference in adjuvant endocrine therapy. Furthermore, our previous study reported that the Arg388 allele of FGFR4 was associated with a poor prognosis for esophageal cancer that was treated with chemoradiotherapy during its early stages (stage I-II), but not during its advanced stages (stage III-IV) [14]. Taken together, these results suggest that FGFR4 could be a crucial component in the early stages of cancer after curative resection or chemoradiotherapy.
Because of the increased need for effective colon cancer adjuvant treatments, we characterized the prognostic role of FGFR4 polymorphism after curative resection in colon cancer patients. The results suggested the molecular mechanism associated with the EMT, which is the rate-limiting step for tissue invasion during colon cancer progression [15].
Materials and Methods1. Patients and samplesThis investigation was conducted to determine the association of genetic polymorphisms and treatment outcomes in colon cancer. The study was approved by the Institutional Review Board of Chonnam National University Hwasun Hospital (CUNH IRB-2014-016). All patients in this study were treated by curative resection for stage III colon adenocarcinoma (American Joint Committee on Cancer, sixth edition) for confirmed adenocarcinoma and gave informed consent for research use of their tissue and blood. Patients who died within 30 days after surgery with postoperative complications were excluded from the study. After surgery, patients received adjuvant chemotherapy based on their performance status or willingness under the current consensus guidelines. Data regarding a patient’s characteristics, history of adjuvant chemotherapy, DFS, and overall survival (OS) were obtained from medical records.
2. Genotyping of FGFR4 in peripheral bloodBlood samples for genotyping were taken before surgery. Genomic DNA was extracted from peripheral blood using a QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA) following the manufacturer’s protocols. Genotyping of the Gly388 allele of FGFR4 was performed by high resolution melting (HRM) analysis using a Rotor Gene 6000 (Corbett Research, Sydney, Australia). Polymerase chain reaction (PCR) primers were as follows: forward 5'-GGAGAGCTTCTGCACAGTGG-3' and reverse 5'-CTTGGCTGTGSTCCTGCT-3'. The reaction mixture for HRM included 200 nM PCR primers, 1 μM SYTO 9 fluorescent dye (Invitrogen, Carlsbad, CA), 0.5 units f-Taq polymerase and 40 ng genomic DNA in a 10 μL reaction volume. The cycling conditions included an initial 5 minutes hold at 95°C, followed by 40 cycles of 95°C for 5 seconds, 65°C for 30 seconds, and 72°C for 20 seconds, with melting temperatures increasing from 78°C to 92°C at 0.1°C/sec. The genotyping results were validated by direct sequencing (ABI PRISM 3100 Genetic Analyzer, Applied Biosystems, Foster City, CA) of 16 samples (6%), and the results were 100% concordant. Appropriate positive/negative and internal controls were included.
1) Microsatellite instability testingThe pentaplex panel of mononucleotide repeats was used for microsatellite instability analysis. This panel is composed of five mononucleotide markers; BAT25, BAT26, NR21, NR22, and NR24. One primer in each pair was labeled with fluorescence (FAM, HEX) at the 5′ end. PCR for all markers was performed in 20 μL reaction volumes with 200 nM PCR primer, 0.5 U f-Taq polymerase, and 50 ng of genomic DNA. The PCR conditions were initial denaturation at 95°C for 5 minutes, followed by 40 cycles of 95°C for 30 seconds, 55°C for 40 seconds and 72°C for 30 seconds, and then final extension at 72°C for 5 minutes. The mixed PCR products with ROX standard were analyzed on an ABI 3130 xl Genetic Analyzer using GeneScan Analysis software (Applied Biosystems).
2) Cell culture and transfectionHuman colorectal cell line HCT 116 was cultured in Dulbecco's modified Eagle medium (HyClone, Logan, UT) supplemented with 10% fetal bovine serum (Gibco BRL, Rockville, MD) and 1% penicillin/streptomycin (Gibco BRL). To generate an FGFR4 overexpressing plasmid, approximately 2.4 kb of a PCR fragment corresponding to the full-length FGFR4 was amplified from HCT 116 cDNA using the following primers: forward 5'-CCCAAGCTTGGAATGCGGCTGCTGCTGGCCCTGTTGG-3' and reverse 5'-CCGCTGGAGTGTCTGCACCCCAGACCCGAAGGGGA-3 ' (underlined sequences are the HindIII and XhoI restriction sites). A pcDNA6-FGFR4-Gly388 plasmid was constructed by cloning the PCR fragment into a pcDNA6 mammalian expression vector, which was verified by restriction endonuclease treatment and DNA sequencing. A mutant FGFR4 cDNA coding for arginine instead of glycine 388, pcDNA6-FGFR4-Arg388, was generated by PCR-mediated sitedirected mutagenesis using the pcDNA6-FGFR4-Gly388 plasmid as a template, two mutagenic primers, and a QuikChange Site-directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s instructions. The mutagenic primers were as follows: forward 5'-GCTGTGCTCCTGCTGCTGGCCAGGCTGTATCG-3' and reverse 5'-GCGCCTGCCCTCGATACAGCCTGGCCAGCAGCAG-3'. The sequence of mutated FGFR4 was verified by DNA sequencing. Transfection was performed using Lipofectamine 2000 (Invitrogen). At 48-hour post-transfection, 5 μg/mL blasticidin (Sigma-Aldrich, St. Louis, MO) was added, and the live cells were selected as stably transfected cells.
3) Cell viability analysisThe cell viability was monitored using a RealTime-Glo MT cell Viability Assay kit (Promega, Madison, WI) following the manufacturer’s instructions.
4) Western blot analysisWhole cell lysates were obtained with radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors (Thermo Fisher Scientific, Waltham, MA). The protein concentrations were measured using a BCA Protein Assay Kit (Pierce, Rockfold, IL). The following antibodies were used: anti-FGFR4, anti-pFRS2α, anti-pSTAT3, anti-pAKT, anti-pERK, and anti-Snail from Cell Signaling Technology (Danvers, MA); anti–E-cadherin from BD Sciences (San Jose, CA); anti-vimentin from Santa Cruz Biotechnology (Santa Cruz, CA); anti–β-actin and anti-Twist from Abcam (Cambridge, UK); anti-CD133 from Miltenyi Biotec (Bergisch Gladbach, Germany); and anti-CD44 from R&D Systems (Minneapolis, MN).
5) Invasion and migration assayThe cell invasion assay was performed using Transwell filter chambers that were coated with 1 μg/mL Matrigel in culture media for 6 hours, then dried at room temperature. The cells were seeded at 2×105 cells in 150 μL medium with 1% bovine serum albumin (BSA) into the upper chamber. Next, 600 μL of medium with 1% BSA and 20 μg/mL fibronectin (Calbiochem, La Jolla, CA) was loaded into the lower chamber. After 24 hours of incubation, cells that invaded to the bottom surface of the Transwell were fixed with 70% ethanol, stained with Diff-Quik solution (Sysmex, Kobe, Japan), and counted in five selected fields. The cell migration was measured using Culture-Inserts (Ibidi, Regensburg, Germany). Briefly, the Culture-Inserts were transferred into 6-well culture plates, after which cells were seeded at a density of 1×105 cell/100 μL in each well of the Culture-Inserts. After 24 hours of incubation, the Culture-Inserts were removed, and cell-free gaps were created. Images of the closed gap were captured at the indicated incubation times.
6) Statistical analysesAssociation analyses between genotypes and clinicopathological characteristics were performed using the chi-squared test and Fisher exact test. Survival curves were calculated using the Kaplan-Meier method, and curves were compared using the log-rank test. The DFS time was calculated from the time of diagnosis of disease to recurrence. The OS time was calculated from the diagnosis of disease to death from any cause, and patients who were alive at the last follow-up were recorded at that time. Univariate analyses were performed using the Kaplan-Meier method and the log-rank test. All variables from univariate analyses with p-values of < 0.1 were incorporated into the multivariate Cox hazard regression model with a stepwise forward procedure. All p-values were derived from a two-tailed statistical test with a 95% confidence interval for evaluation of the statistical significance between groups. All statistical analyses were performed using SPSS statistical software for Windows ver. 21.0 (IBM Corp., Armonk, NY), and a p < 0.05 was considered to indicate significance.
Results1. Study populationA total of 324 consecutive patients resected for stage III colon cancer between May 2004 and December 2011 were reviewed. Among the patients, 273 who met the inclusion criteria were enrolled in this study. Their median age was 70 years (range, 34 to 85 years). After surgery, 127 (47%) patients received 5-FU and leucovorin (FL) and 123 (45%) received a combination of folinic acid, 5-FU, and oxaliplatin (FOLFOX) as an adjuvant chemotherapy; 23 patients (8%) did not receive any adjuvant chemotherapy (Table 1). During follow-up (median, 41 months), 66 patients experienced disease recurrence, while 41 died from colon cancer. The rates of the 3-year DFS and 5-year OS were 74.2% (95% confidence interval [CI], 68.71 to 79.69) and 80.9% (95% CI, 75.22 to 85.58), respectively.
2. Incidence of FGFR4 Gly388Arg polymorphismOut of 273 patients, 92 (34%) were homozygous for the Gly388 allele (GG), 146 were heterozygous (GA, 53%), and 35 were homozygous (AA, 13%) for the Arg388 allele. The percentage of patients with the Arg388 allele was higher than in some previous reports (50%-60%) [8], but consistent with another study reporting an incidence of 11.8% in colorectal cancer patients [9]. No significant association was found between clinical or histopathological tumor characteristics and the FGFR4 genotype (Table 1).
3. Treatment outcomes according to FGFR4 genotypeThere was no significant difference in the DFS between patients heterozygous for the Gly388 and Arg388 alleles (p=0.500). However, the OS was significantly better in patients homozygous for the Gly388 allele than for heterozygous patients or patients with the Arg388 allele (p=0.017). Based on these results, further analyses were performed on the two groups of patients with the Gly388 allele and the Arg388 carrier patients (heterozygous and Arg388 allele) (Fig. 1). Using univariate analyses, N stage, perineural invasion (PNI), and tumor differentiation, the patients receiving adjuvant chemotherapy were significantly associated with the DFS. The OS, age, N stage, tumor differentiation, lymphocyte response, adjuvant chemotherapy and FGFR4 genotype were significantly associated with prognosis (Table 2). Multivariate analyses showed that the N stage (N2 vs. N1), presence of PNI, and absence of adjuvant chemotherapy were significant independent factors for the DFS. In addition, age (≥ 70 years), N stage (N2 vs. N1), poor differentiation, absence of a lymphocyte response, absence of adjuvant chemotherapy, and Arg388 carriers were significantly associated with poor prognoses for the OS (Table 3).
4. The Gly388 allele of FGFR4 was an indicator of a good prognosis, regardless of adjuvant chemotherapyAs previously mentioned, the OS was significantly higher in patients with the Gly388 allele of FGFR4 than with Arg388 carriers. To evaluate the effects of adjuvant regimen, we analyzed the OS according to genotype, stratified by adjuvant chemotherapy. In the group without treatment, the 5-year OS percentages in patients with the Gly388 allele and Arg388 carriers were 88.9% and 30.0%, respectively (p=0.025). However, the difference in the 5-year OS percentages according to genotypes was attenuated in the FL-treated group compared with the no treatment group (p=0.06), and was comparable to the FOLFOX-treated group in patients with the Gly388 allele and Arg388 carrier patients (p=0.174). These results suggested that intensified adjuvant chemotherapy could overcome the poor prognosis of patients with the Arg388 allele in a similar manner to patients with the Gly388 allele. DFS and OS were analyzed according to adjuvant chemotherapy, stratified by the FGFR4 genotype, to determine the possible benefits of adjuvant chemotherapy for each genotype. Regarding the DFS and OS for Arg388 carriers, the FOLFOX-treated group had the most significant improvements when compared to the group without treatment. However, there was no significant difference in survival outcomes from the adjuvant chemotherapy regimen for Gly388 carriers with a good prognosis (Fig. 2).
5. The overexpression of the Arg388 and Gly388 alleles of FGFR4 promoted colon cancer cell proliferationTo determine if the presence of the Arg388 allele was associated with colorectal cancer cell progression, we conducted FGFR4 genotype analysis with cDNAs from eight colorectal cancer cell lines. The homozygous Gly388 allele was present in HCT 116 and SW480 cells, the heterozygous Gly388 allele was present in HT29, CaCo2, and KM12c cells, and the homozygous Arg388 allele was present in DLD1, DKO1, and HCT15 cells. Because the heterozygous and Arg388 alleles had poorer survival rates than the Gly388 allele, we selected the HCT 116 cell line, which did not contain the Arg388 polymorphism, to overexpress the Arg388 allele of FGFR4. The Arg388 and Gly388 stable expression plasmids were constructed and transfected into HCT 116 cells. An empty vector was also transfected into control cells. The overexpression of Arg388 and Gly388 of FGFR4 was confirmed by western blotting.
To determine the effects of Arg388 and Gly388 overexpression on downstream signaling, we analyzed the expression levels of FGFR4 downstream targets by western blotting. Phosphorylation of the primary FGFR target, FRS2α, was increased in overexpressed Arg388 and Gly388 cells compared with control cells. Although FRS2α was increased in both transfected cells, FRS2α was increased more in overexpressed Arg388 than in overexpressed Gly388 cells. Furthermore, the expression of downstream signals of FGFR4, including of pSTAT3, pAKT, and pERK, was also further increased in overexpressed Arg388 than Gly388 cells (Fig. 3A).
The 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide (MTT) assay was performed to assess the cell proliferation of overexpressed Arg388 and Gly388 cells. The cell growth rates increased in overexpressed Gly388 and Arg388 cells when compared with the control cells, but the growth rate of overexpressed Arg388 cells was similar to that of overexpressed Gly388 cells. These results indicated that FGFR4 is associated with increasing cell proliferation, but that there was no difference between cells overexpressing Gly388 and Arg388 (Fig. 3B).
6. The Arg388 allele of FGFR4 induced EMT signals and enhanced invasion and migrationThe EMT involves profound changes in cell morphology and behavior. This process plays a crucial role in the early stages of cancer recurrence and metastasis. To study the role of the genotype in the EMT, we assessed the roles of the Arg388 allele compared with the Gly388 allele of FGFR4 during induction of EMT changes in stably transfected cells. Using western blot analyses, E-cadherin was significantly reduced in Arg388 overexpressed cells when compared with Gly388 overexpressed and control cells. In addition, there was increased expression of vimentin and Twist in Arg388 overexpressed cells compared with Gly388 overexpressed and control cells. These results suggested that the Arg388 allele of FGFR4 induces the EMT process (Fig. 4A). To investigate the functional properties of the EMT inducers, we conducted cell invasion and migration studies of FGFR4 transfected cells. Stably transfected Arg388 Gly388 cells, including the control HCT 116 cell, were seeded in Transwell filter chambers, and the cell motility towards human plasma fibronectin was determined. When compared with control cells, the Gly388- and Arg388-transfected cells showed significantly more cell invasion. Moreover, the Arg388-transfected cells were more invasive than the Gly388-transfected cells (Fig. 4B). Similar results were also seen using a wound healing assay (Fig. 4C). Together, these results suggested that both the Arg388-transfected and Gly388-transfected cells significantly increased the migration and invasion in colorectal cancer cells, but that overexpression of the Arg388 allele was more robust.
DiscussionThe percentage of patients with recurrent or metastatic colon cancer who are cured is less than 10%; therefore, there have been extensive studies conducted to improve these prognoses. To improve adjuvant treatments, actual high-risk patients must be identified and adjuvant chemotherapies tailored to prevent recurrence developed. Therefore, the identification of biomarkers to select optimal patients and druggable targets of colon cancer would increase the efficacies of current treatments greatly.
Previous studies to define the functional role of FGFR4 have mainly characterized the overexpression of FGFR4 in cancer model systems. Recently, FGFR4 polymorphisms have been evaluated in several tumors, and the characteristics of the Arg388 allele of FGFR4 have been compared with the Gly388 allele, showing similar features of overexpression of FGFR4 [16,17]. In addition to in vitro studies, the Arg388 allele of FGFR4 has been associated with a poor prognosis after surgery in prostate and breast cancer patients [6,17]. It has been suggested that FGFR4 polymorphism can be a surrogate marker to reflect the abnormal FGFR4 pathway in tumors.
The results of the present study showed that the FGFR4 genotype has the highest odds ratio in the prediction of the OS. Contrary to previous studies of FGFR4 polymorphism using various tumor stages and treatment populations [6,7,11], the present study only characterized stage III colon cancer patients to clearly define the role of FGFR4 polymorphism in the OS after curative resection. Moreover, our results determined the role of adjuvant chemotherapy according to genotypes. Patients with the Gly388 allele of FGFR4 had a good prognosis, regardless of adjuvant chemotherapy. However, Arg388 allele carrier patients had a poor prognosis, although their OS was improved by treatment with an intensified chemotherapy regimen (FOLFOX > FL > no treatment). Based on these results, the Arg388 carriers are candidates for adjuvant chemotherapy, while patients with the Gly388 allele of FGFR4 are not, even though they have stage III colon cancer. Given the lack of effective biomarkers for colon cancer in an adjuvant setting, our results suggest that the FGFR4 genotype can be used to identify optimal treatment strategies and develop new treatment targets.
The FGFR4 genotype’s mechanism of action for aggressive tumors behavior reportedly involves receptor stability rather than a difference in protein expression [6,7]. In the prostatic cancer model, the presence of the Arg388 allele of FGFR4 increased receptor stability and sustained receptor activation following ligand binding when compared with the Gly388 allele. These changes were the result of increased activity of the SRF, AP, and ERK pathways [12,17]. A recent study reported that functional changes in FGFR4 polymorphisms originated from substitution of the conserved glycine 388 residue to a charged arginine residue that altered the transmembrane spanning segment and exposed a membrane proximal STAT3 binding site [18]. These results explained the functional changes resulting from the genetic polymorphism during cancer progression. Therefore, genetic studies should characterize heterogeneous somatic mutations of tumors and genetic variation in patients to identify therapeutic targets.
To establish the biologic significance of FGFR4 polymorphism in colon cancer, we first evaluated protein expression according to various genotypes in colon cancer cell lines derived from patients and human tumor samples. Overall, the level of FGFR4 expression was not significantly different in each genotype (unpublished data). These results are similar to those of previous studies of breast and gastric cancers that reported no significant correlation with FGFR4 genotype and FGFR4 protein or mRNA expression [6,19]. Although the association of FGFR4 genotype and protein expression was not conclusive, gene polymorphism is an easier and more reproducible method than immunohistochemical staining because of the variability in expression from tumor heterogeneity or staining methods. Therefore, FGFR4 polymorphism could be a useful surrogate biomarker of FGFR4 function in cancer patients.
To validate the function of the FGFR4 genotype in the recurrence of cancer, its effects on the EMT were evaluated in transfected cell lines. The EMT is an early event that facilitates infiltration of surrounding tissue, ultimately resulting in metastasis to distant sites [20]. Several studies have reported that STAT3 may play an important role in establishing cell polarity during directed cell migration, which is essential to EMT and carcinoma metastasis using the insulin like growth factor-1 or the interleukin 1/STAT3 pathway [21,22]. Therefore, the present study was conducted to characterize induction of the EMT signal via STAT3 and pFRS2 activation according to the colon cancer genotype. Notably, the Arg388 allele of FGFR4 resulted in a stronger activation of pFRS2 and STAT3, which are known as downstream signals for FGFR4, as well as in stronger activation of EMT-associated proteins, such as vimentin, Twist, and the loss of E-cadherin, when compared with the Gly388 allele of FGFR4. We also showed that the Arg388 allele of FGFR4 had more invasive and migratory activities than the Gly388 allele. To date, few reports have associated the FGFR4 genotype with the STAT3 and EMT pathway during tumorigenesis. Our results support the use of FGFR4 as a therapeutic target in adjuvant treatment for colon cancer.
Small molecular inhibitors targeting the FGFR, especially FGFR1-3, have recently been developed for cancer therapy, including AZD4547, BGJ398, and dovitinib [23-25]. Although pan-FGFR inhibitors have also been developed [26-28], the exact mechanisms of action involving tumors are not fully understood, and it is still difficult to select optimal patients who can benefit from these agents. Furthermore, kinase inhibitors for multiple tyrosines can block many signal pathways and cause significant side effects involving their toxicities. Pan-EGFR inhibitors can induce dose-limiting toxicities in the clinic because of potent inhibition of FGFR1 and FGFR3, resulting in soft tissue mineralization and hyperphosphatemia [29]. To circumvent these problems, a close examination of the specific function of each type of FGFR is necessary. BLU9931 is the first selective FGFR4 inhibitor that achieved improvement of hepatocellular carcinoma prognosis [30]. In hepatocellular carcinoma, there is a well known association of FGFR4 with its β-klotho co-receptor and expression of its ligand, FGF19. However, there has been no report of a specific FGFR4 pathway inhibitor for colon cancer patients. Accordingly, further studies to validate FGFR4 polymorphism as a biomarker are needed in a larger population of colon cancer patients, including patients with stage II colon cancer. In addition, future studies should be directed toward the development of an effective FGFR4 pathway inhibitor or STAT3 inhibitor.
ConclusionThe present study employed FGFR4 polymorphism to help identify treatments for high risk patients with stage III colon cancer. By inhibiting the EMT pathway, FGFR4 can also be used as a new therapeutic target in adjuvant treatment. Based on these findings, the targeted therapy should involve both tumor-specific biology and the stage of the disease, and FGFR4 inhibitors could be used in clinical investigations to assess its efficacy on a subset of colon cancer patients according to their FGFR4 polymorphisms.
AcknowledgmentsThis research was supported by the National Research Foundation of Korea grant (MRC, 2011-0030132) funded by the Korea government (MSIP). The biospecimens and data used for this study were provided by the Biobank of Chonnam National University Hwasun Hospital, a member of the Korea Biobank Network. We thank Ji-Na Choi and Mi-Ra Park for conducting the experimental studies.
Fig. 2.(A-D) Survival outcomes after adjuvant chemotherapy according to FGFR4 genotype. FGFR4, fibroblast growth factor receptor 4; FL, fluorouracil and leucovorin; FOLFOX, combination of folinic acid, fluorouracil, and oxaliplatin. 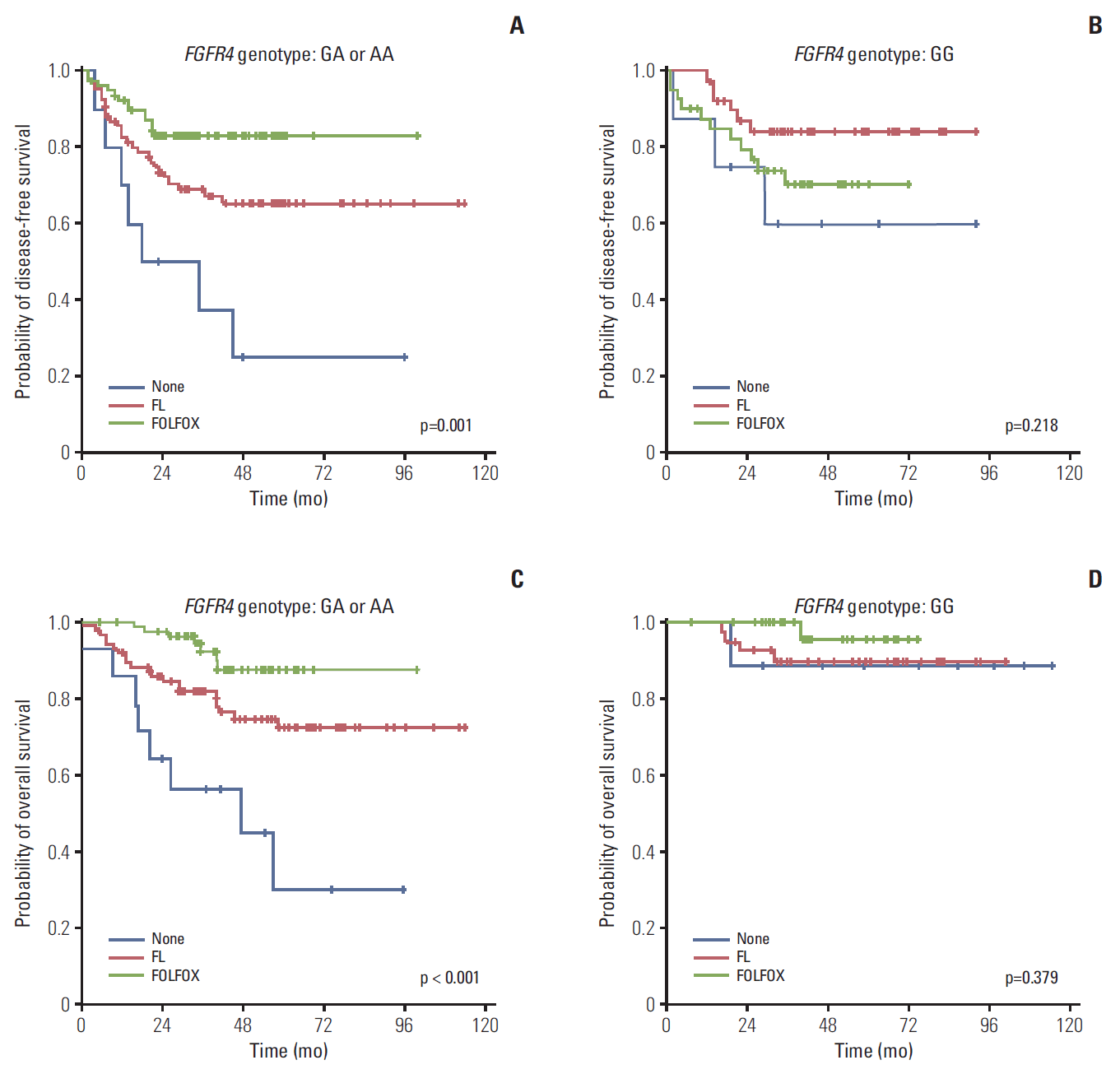
Fig. 3.Fibroblast growth factor receptor 4 (FGFR4) dependent downstream signal and cell proliferation. (A) The expression of downstream signals of FGFR4, including pSTAT3, pAKT, and pERK, were increased more in overexpressed Arg388 than in overexpressed Gly388 cells. (B) However, the growth rate of overexpressed Arg388 cells was similar to that of overexpressed Gly388 cells. *p < 0.01 compared with vector control. 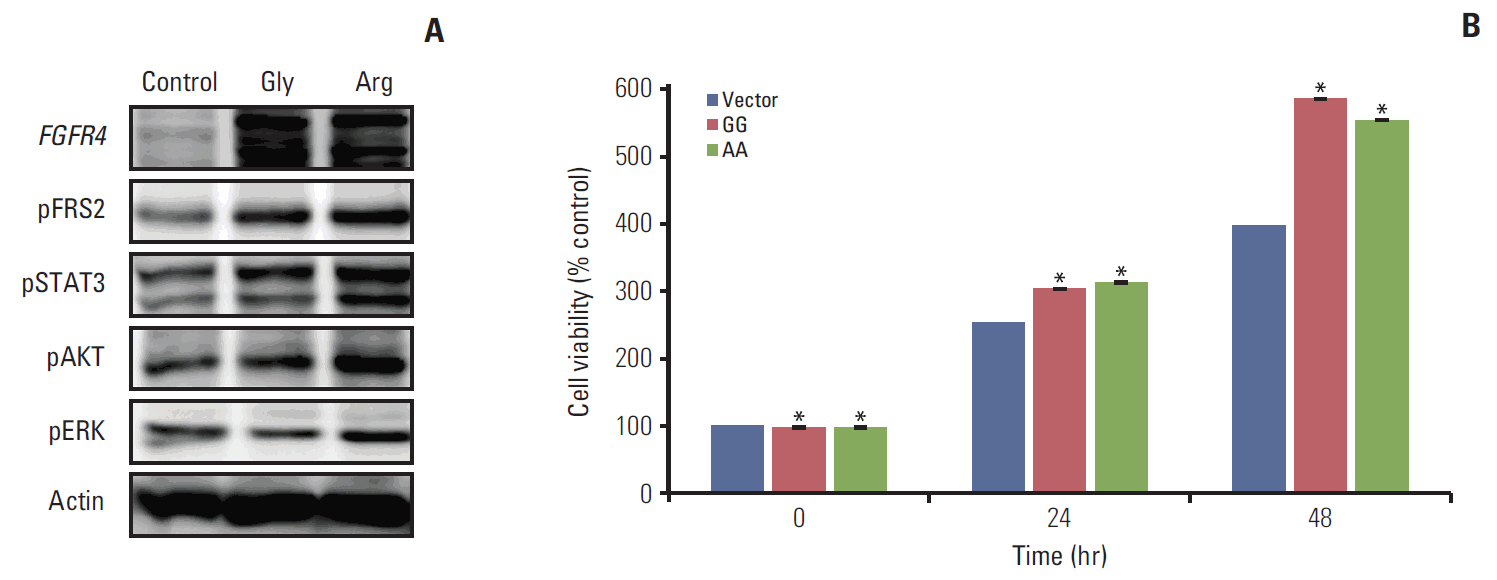
Fig. 4.Effect of fibroblast growth factor receptor 4 (FGFR4) genoypte on epithelial-mesenchymal transition (EMT) signals, invasion, and migraion. (A) The Arg388 allele of FGFR4 induces the EMT markers including, vimentin and Twist, while it decreased E-cadherin. (B) When compared with control cells, the Gly388- and Arg388-transfected cells showed significantly more cell invasion. In addition, Arg388-transfected cells were more invasive than Gly388-transfected cells. (C) Similar results were also seen using a wound healing assay. *p < 0.01. 
Table 1.Patient and clinicopatholgic characteristics
Table 2.Univariate analysis for survival Table 3.Multivariate analysis for survival References1. Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–29.
2. Burke D, Wilkes D, Blundell TL, Malcolm S. Fibroblast growth factor receptors: lessons from the genes. Trends Biochem Sci. 1998;23:59–62.
3. Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18:1855–62.
4. Dieci MV, Arnedos M, Andre F, Soria JC. Fibroblast growth factor receptor inhibitors as a cancer treatment: from a biologic rationale to medical perspectives. Cancer Discov. 2013;3:264–79.
5. Spinola M, Leoni V, Pignatiello C, Conti B, Ravagnani F, Pastorino U, et al. Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J Clin Oncol. 2005;23:7307–11.
6. Thussbas C, Nahrig J, Streit S, Bange J, Kriner M, Kates R, et al. FGFR4 Arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J Clin Oncol. 2006;24:3747–55.
7. Shen YY, Lu YC, Shen DP, Liu YJ, Su XY, Zhu GS, et al. Fibroblast growth factor receptor 4 Gly388Arg polymorphism in Chinese gastric cancer patients. World J Gastroenterol. 2013;19:4568–75.
8. Liu R, Li J, Xie K, Zhang T, Lei Y, Chen Y, et al. FGFR4 promotes stroma-induced epithelial-to-mesenchymal transition in colorectal cancer. Cancer Res. 2013;73:5926–35.
9. Heinzle C, Gsur A, Hunjadi M, Erdem Z, Gauglhofer C, Stattner S, et al. Differential effects of polymorphic alleles of FGF receptor 4 on colon cancer growth and metastasis. Cancer Res. 2012;72:5767–77.
10. Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, Schmitt M, et al. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 2002;62:840–7.
11. Spinola M, Leoni VP, Tanuma J, Pettinicchio A, Frattini M, Signoroni S, et al. FGFR4 Gly388Arg polymorphism and prognosis of breast and colorectal cancer. Oncol Rep. 2005;14:415–9.
12. Wang J, Yu W, Cai Y, Ren C, Ittmann MM. Altered fibroblast growth factor receptor 4 stability promotes prostate cancer progression. Neoplasia. 2008;10:847–56.
13. Streit S, Bange J, Fichtner A, Ihrler S, Issing W, Ullrich A. Involvement of the FGFR4 Arg388 allele in head and neck squamous cell carcinoma. Int J Cancer. 2004;111:213–7.
14. Shim HJ, Shin MH, Kim HN, Kim JH, Hwang JE, Bae WK, et al. The prognostic significance of FGFR4 Gly388 polymorphism in esophageal squamous cell carcinoma after concurrent chemoradiotherapy. Cancer Res Treat. 2016;48:71–9.
15. Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11–9.
16. Desnoyers LR, Pai R, Ferrando RE, Hotzel K, Le T, Ross J, et al. Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene. 2008;27:85–97.
17. Yu W, Feng S, Dakhova O, Creighton CJ, Cai Y, Wang J, et al. FGFR-4 Arg388 enhances prostate cancer progression via extracellular signal-related kinase and serum response factor signaling. Clin Cancer Res. 2011;17:4355–66.
18. Ulaganathan VK, Sperl B, Rapp UR, Ullrich A. Germline variant FGFR4 p.G388R exposes a membrane-proximal STAT3 binding site. Nature. 2015;528:570–4.
19. Ye Y, Shi Y, Zhou Y, Du C, Wang C, Zhan H, et al. The fibroblast growth factor receptor-4 Arg388 allele is associated with gastric cancer progression. Ann Surg Oncol. 2010;17:3354–61.
20. Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelialmesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–81.
21. Sanchez-Lopez E, Flashner-Abramson E, Shalapour S, Zhong Z, Taniguchi K, Levitzki A, et al. Targeting colorectal cancer via its microenvironment by inhibiting IGF-1 receptor-insulin receptor substrate and STAT3 signaling. Oncogene. 2016;35:2634–44.
22. Balanis N, Wendt MK, Schiemann BJ, Wang Z, Schiemann WP, Carlin CR. Epithelial to mesenchymal transition promotes breast cancer progression via a fibronectin-dependent STAT3 signaling pathway. J Biol Chem. 2013;288:17954–67.
23. Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72:2045–56.
24. Guagnano V, Kauffmann A, Wohrle S, Stamm C, Ito M, Barys L, et al. FGFR genetic alterations predict for sensitivity to NVPBGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2012;2:1118–33.
25. Lee SH, Lopes de Menezes D, Vora J, Harris A, Ye H, Nordahl L, et al. In vivo target modulation and biological activity of CHIR-258, a multitargeted growth factor receptor kinase inhibitor, in colon cancer models. Clin Cancer Res. 2005;11:3633–41.
26. Tan L, Wang J, Tanizaki J, Huang Z, Aref AR, Rusan M, et al. Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc Natl Acad Sci U S A. 2014;111:E4869–77.
27. Tabernero J, Bahleda R, Dienstmann R, Infante JR, Mita A, Italiano A, et al. Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2015;33:3401–8.
28. Zhao G, Li WY, Chen D, Henry JR, Li HY, Chen Z, et al. A novel, selective inhibitor of fibroblast growth factor receptors that shows a potent broad spectrum of antitumor activity in several tumor xenograft models. Mol Cancer Ther. 2011;10:2200–10.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||