Introduction
Epstein-Barr virus (EBV) has been implicated in the development of a wide range of cancers such as nasopharyngeal carcinoma (NPC), Hodgkin's disease, and Burkitt's lymphoma since its discovery as the first human tumor virus [1]. EBV encodes multiple viral proteins, and the expression patterns of those proteins in human malignancies are variable. Some of viral products such as latent membrane protein (LMP)-1 and EBV nuclear antigen 2 have been suggested to be important in the oncogenic process of many EBV-associated cancers.
EBV is found in approximately 10% of gastric cancers (GCs) [2], and is proposed to have a critical role in EBV-associated GCs since the virus is present all cancer cells and carcinoma is formed by the proliferation of a single EBV infected cell [3]. However, the specific genes from EBV genome contributing to the carcinogenesis of GCs still remain unknown. The latency pattern in EBV-carrying GCs is distinct from the other EBV-associated malignancies by the absence of LMP 1, LMP2B [4] and the expression of the BARF1 [5]. Some EBV genes are suggested to have an oncogenic role in EBV-carrying GCs. For example, BARF1, which is exclusively expressed in EBV-associated GCs, has oncogenic properties in many cell types such as monkey kidney epithelial cell line [6] and human B lymphocytes [7], and human epithelial cell line HBE [8]. In addition, cyclin D1 induced in BARF1-transfected epithelial cells was overexpressed in EBV-associated GCs, indicating an interaction of viral BARF1 and cyclin D1 [9]. However, BARF1 transcripts have never been shown to produce protein in EBV-positive GCs. Thus, it requires further study to confirm the oncogenic role of BARF1 in EBV-associated GCs.
As EBV-infected GC cells show the specific latency pattern of viral gene expression and usually do not express the major oncogenic LMP1, we tried to identify the specific transcripts implicated in the carcinogenesis of EBV-positive GCs from EBV-infected gastric cancer cell lines. In this study, we searched for EBV-associated clones in cDNA libraries established from the EBV-infected GC cell line, SNU719, to find novel viral oncogenes in GCs, and investigated their expression in SNU719 cells.
Materials and Methods
1. Cell line
The SNU719 cell line was used. It is a naturally derived EBV-infected cell line, which was established from a 53-year-old male patient with primary stomach cancer tissue in July 1991 [10]. It was obtained from the Korean Cell Line Bank (Seoul, Korea) and was cultured in RPMI-1640 supplemented with 10% heat-inactivated FBS (Hyclone, Logan, UT) at 37℃ in a humidified CO2 incubator.
2. Bioinformatic analysis of EBV-associated expressed sequence tags (ESTs)
Two cDNA libraries derived from SNU719 are available in the National Center for Biotechnology Information (NCBI) UniGene database (http://www.ncbi.nlm.nih.gov/unigene). The individual ESTs were searched against the mRNA subset extracted from the GenBank data base. The selected clones were obtained from 21C frontier Gene Bank (Daejeon, Korea). Plasmid DNAs were extracted and sequencing reactions were performed using a BigDye Terminator Sequencing kit (Applied Biosystems, Foster City, CA). After the unincorporated dye terminator was removed from the sequencing reaction, the reaction products were applied to genetic analyzers (Applied Biosystems).
3. Reverse transcriptase polymerase chain reaction (RT-PCR)
After cells were partitioned into nuclear and cytoplasmic fraction, RNA was extracted by PARIS (Ambion, Austin, TX). Five hundred nanograms of RNA were used to synthesize cDNA using a random hexamers. Reverse transcription was carried out with Superscript Reverse Transcriptase (Invitrogen, Carlsbad, CA). PCR was performed with primers which were used in the previous studies [11,12]. The PCR reaction mixture consisted of 1× PCR premix (Takara, Shiga, Japan), 5 pmol of each primer, and 1 µL of cDNA. PCR involved 25 cycles of 95℃, 30 seconds; 50-60℃, 30 seconds; 72℃, 1 minute followed by 10 minutes at 72℃ to complete reaction. This experiment was repeated three times.
Results
1. Analysis of EBV-associated ESTs from the SNU719 cDNA library
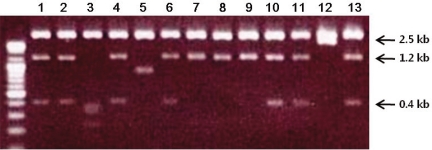
ESTs are useful for the differential and quantitative analysis for the evaluation of gene expression profiles in a given tissue. We found two SNU719 cDNA libraries (S7SNU719 and S7SNU719S1) established from SNU719, EBV-infected gastric cancer cell line in NCBI's UniGene database. The total number of ESTs from the two libraries was 4,701. The individual EST was searched against the mRNA subset extracted from the GenBank data base. We selected 13 clones that had a possibility of being transcribed from the EBV genome by Blast analysis. After each clone was linearized with EcoR I and Not I, it was electrophoresed in 1% agarose gel (Fig. 1). We classified them into five groups based on size. Sequencing the clone from each five groups revealed that three groups of clones-1.2 kb, 1.6 kb, and 2.5 kb-were derived from EBV genome and were related with the RPMS1 transcript that spans the whole BamHI-A rightward transcript (BART) region of EBV (Table 1). BART transcripts are a group of various splicing variants transcribed from 138352 to 160531 on the wild-type EBV genome, and some open reading frames (ORFs) in those BART cDNAs have been actively investigated such as RPMS1 and A73. Through BLAST sequence alignment, three groups of clones were included in RPMS1 cDNA. Group 3 clone covered most of the ORF of RPMS1, whereas Group 1 and 2 clones showed no overlap with any ORFs of RPMS1 and A73 (Fig. 2).
2. Analysis of cytoplasmic and nuclear EBV transcripts from EBV-infected gastric cancer cell lines
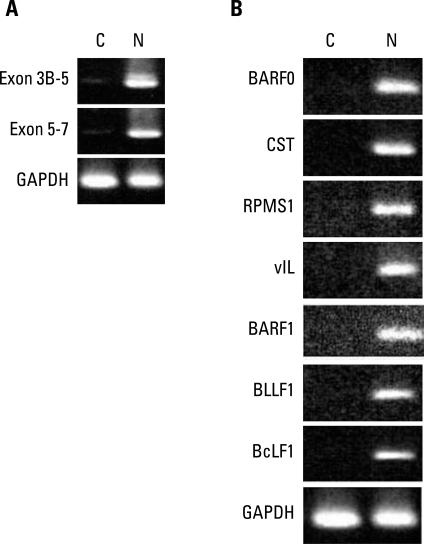
Some BARTs such as RPMS1 and A73 have ORFs, and they have been shown to produce proteins in vitro with significant biologic functions relevant to carcinogenesis. However, it has never been demonstrated in naturally EBV infected cells. Since we identified the various BARTs in EBV-associated GC, we decided to investigate whether or not they might be acting as mRNAs. If gene transcripts act as mRNAs, they will be detected in the cytoplasm. Therefore, we probed for BART RNAs of SNU719 in the cytoplasm. After cells were partitioned into nuclear and cytoplasmic fractions, RNA was extracted and RT-PCR was performed with primers specific for the spliced exons 3B-5 and 5-7 of BART. BART RNA was almost completely in the nucleus (Fig. 3A). Some latent and lytic transcripts that are expressed in EBV-associated GCs were also detected in the nucleus only (Fig. 3B). However, we were not able to confirm the expression of BART transcripts by Northern blot analysis. Although we did not detect any BART transcripts in the cytoplasm of SNU719 cells, it is still possible that BART transcripts might have been translated into protein that functioned in the development of GCs. This role would likely have been in the early stage of tumorigenesis, but not in fully developed cancer cells. We are planning to try RNA in situ hybridization for paraffin embedded tissues to further explore the cytosolic BART transcripts in dysplastic cells triggered by EBV infection, which is expected to clarify the existence of BARTs in many different stages of gastric epithelial cells undergoing transformation.
Discussion
BART RNAs were originally identified by analysis of the cDNA libraries established from the NPC cell line [13]. Since then, they have been demonstrated to be expressed in many EBV-associated malignancies including EBV-positive GCs. In this study, we identified some BART RNAs in the EBV-positive GC cell line. It is notable that all EBV-related clones that we found from the cDNA library of EBV-positive GC cell line turned out to be related with BART, which suggests that BART RNAs are quite possibly the predominant transcripts from EBV genome in EBV-infected GCs.
Among these clones, the largest one (2.5 kb) contained the ORF of RPMS1 indicating that RPMS1 protein might be translated if that cDNA acts as mRNA. In fact, RPMS1 already has been shown to produce a nuclear protein in vitro, which could be detected using specific antibodies that recognize artificially expressed RPMS1 protein. These findings strongly support the possibility that RPMS1 could act as mRNA in vivo. However, we observed RPMS1 transcripts exclusively expressed in the nucleus of EBV-positive GC cells by performing RT-PCR after nuclear/cytoplasmic fractionation, which argues against them acting as mRNAs. Furthermore, previous studies also failed to detect the endogenous RPMS1 protein in NPC or Burkitt lymphoma cases with the specific monoclonal antibodies that were verified to be able to detect the recombinant RPMS1 protein [11,14,15]. Thus, it still remains uncertain that RPMS1 transcripts produce a detectable protein in natural EBV infections.
Despite the lack of evidence of its acting as mRNA, RPMS1 clearly has some biologic functions that are quite relevant to tumorigenesis. Artificially expressed RPMS1 protein antagonizes the transcription activation by Notch 1 or EBVA2 through its interaction with the CBF1-associated corepressor complex [15], and enhances anchorage-independent growth of 293 cells and produce tumors in nude mice [16]. A73, another BARTs-derived potential protein, has been translated in vitro and was shown to bind to the cellular RACK1 and regulate calcium influx, even though the existence of endogenous A73 protein has never been demonstrated [11]. These biologic functions could fit well with a pathologic role of EBV in EBV-related cancers. Therefore, the possibility still remains, and we can speculate that RPMS1 might be transiently expressed in specific circumstances only and contribute to the development of cancer. For example, RPMS1 might be translated into the protein under a certain precancerous stage, but not fully-developed cancer cells. Thus, it could be meaningful to try to investigate the presence of RPMS1 protein in non-cancer cells that are only infected by EBV or cells that are show dysplastic changes after EBV infection.
The abundant BART RNA transcripts have been implicated in carcinogenesis by contributing to the production of miRNAs. Since the first discovery of EBV miRNAs from the Burkitt's lymphoma cells, over 20 miRNAs have been identified [17,18]. These miRNAs were shown to be present in EBV-infected GC cell lines and the tumor tissues from patients [19]. Some targets for miRNAs have been unraveled such as EBV DNA polymerase [20], EBV LMP1 [21], and p53 up-regulated modulator of apoptosis [22]. Interestingly, these miRNAs are derived mainly from the introns prior to splicing of the BART primary transcripts, and their expression is largely predicted by the level of expression of the BART RNAs [18]. Based on the close correlation of expression of BART transcripts and BART miRNAs, it can be suggested that the primary goal of the transcription of BART might be to promote the production of miRNAs, not to produce the viral proteins. Alternatively, BART transcripts might have dual functions, serving as oncogenic proteins in precancerous stage and then supplying miRNAs to support cell survival or immune evasion in established cancer cells.
Conclusion
In this study, we showed that BART transcripts are the predominant viral transcripts expressed in EBV-associated GCs. We also observed they are located in the nucleus only, indicating that BART transcripts are less likely to produce functional proteins than they are to play a role in carcinogenesis of EBV-associated GCs.