AbstractPurposePaternally expressed gene 10 (PEG10), first identified as an imprinted gene, is paternally expressed and maternally silenced. In hepatocellular carcinoma (HCC), PEG10 has been identified as a potential target gene located within the amplified 7q21 locus. The purpose of this study was to investigate the expression of PEG10 protein in HCC and evaluate its prognostic significance.
Materials and MethodsPEG10 protein expression was examined by immunohistochemistry in tumor tissues from 218 HCC patients undergoing curative resection. Furthermore, the relationships between PEG10 expression and clinicopathologic features or postoperative survival of HCC patients were evaluated. The median follow-up period was 119.8 months for survivors.
ResultsPEG10 expression was observed in 148 of the 218 HCCs (67.9%) and was significantly correlated with younger age, female, higher Edmondson grade, microvascular invasion, intrahepatic metastasis, higher American Joint Committee on Cancer T-stage, and higher α-fetoprotein level. PEG10 expression was an independent predictor of early recurrence (p=0.013), and it showed an unfavorable influence on recurrence-free survival (p < 0.001). A subgroup analysis showed that among patients with α-fetoprotein ≤ 20 ng/mL (80 patients), the PEG10-positive group also showed an unfavorable influence on recurrence-free survival (p=0.002). Moreover, a multivariate survival analysis identified PEG10 as an independent predictor of shorter recurrence-free survival (p=0.005). PEG10 expression showed an unfavorable influence on overall survival (p=0.007) but was not an independent predictor of shorter overall survival (p=0.128).
IntroductionThe overall prognosis of hepatocellular carcinoma (HCC) patients remains poor due to many risk factors, including late diagnosis, high incidence of tumor recurrence after curative resection, and resistance to chemotherapy [1,2]. It is a necessity to identify patients with poor prognosis for timely intervention. Since cancer is a heterogeneous disease that encompasses a wide variety of cell types, effective therapeutic approaches require a better understanding of molecular mechanisms that drive oncogenesis and clarify the responsiveness of different cancer phenotypes to potential therapeutic agents [3].
Paternally expressed gene 10 (PEG10) was first described by Ono et al. [4] as an imprinted gene with an active paternal allele but silent maternal allele and is generally not expressed in the normal adult liver. It has been suggested that PEG10 is derived from a retrotransposon that was previously integrated into the mammalian genome [4]. PEG10 was overexpressed in a variety of human cancers, including leukemia, breast cancer, prostate cancer, pancreas cancer, gallbladder cancer, and HCC [5-7]. Furthermore, PEG10 was a poor-prognosis related biomarker for gallbladder adenocarcinoma [6]. In HCC, PEG10 has been identified as a potential target gene located within the amplified 7q21 locus [8-10]. A functional role for PEG10 in the growth-promoting activities has been demonstrated in the HCC cells [7]. Ip et al. [8] reported a significant progressive trend of increasing PEG10 expression from the putative pre-malignant adjacent livers to early resectable HCCs, and to late inoperable HCCs. However, the prognostic significance of PEG10 protein expression in HCC has not yet been elucidated.
In this study, we investigated PEG10 protein expression by immunohistochemistry in order to identify a marker associated with tumor recurrence and to evaluate the prognostic significance of PEG10 in 218 HCC patients with longterm follow-up.
Materials and Methods1. Tissue samplesLiver tissue samples from 218 surgically resected HCC were collected between July 2000 and May 2006 from Samsung Medical Center, Seoul, Korea. The cases were selected based on the following criteria: pathological diagnosis of HCC and curative resection of tumor without preoperative or postoperative adjuvant therapy. Curative resection was defined by a complete resection of all tumor nodules with clear microscopic resection margins and no residual tumors as indicated by a computed tomography scan one month after surgery. Tumor differentiation was determined based on the criteria proposed by Edmondson and Steiner [11]. A microvascular invasion was considered present when at least one or more endothelial cells or tunica media of the vessel surrounded the neoplastic cell group. Intrahepatic metastasis and multicentric occurrence were defined in accordance to the previously reported criteria [12]. All patients were staged using the American Joint Committee on Cancer (AJCC) staging system [13] and Barcelona Clinic Liver Cancer (BCLC) staging classification [14]. Early recurrence after curative resection of HCC was defined as an intrahepatic or extrahepatic recurrence within 2 years [15]. The Institutional Review Board of Samsung Medical Center granted approval for this study.
Patients were followed every 3 months after surgery. Tumor recurrence was detected by three phase dynamic computed tomography scans or a magnetic resonance imaging. The median follow-up period was 119.8 months (range, 14.0 to 151.4 months) for survivors and the follow-up period for recurrence was at least 24 months. Recurrence-free survival (RFS) was defined as from the date of the operation until the detection of tumor recurrence.
Two pathologists reviewed the slides to identify the representative tumor areas and marked the formalin-fixed paraffin-embedded blocks. Two tissue cores with a diameter of 2.0 mm were punched from the marked areas of the donor tissue block and re-embedded into the recipient paraffin blocks at defined positions. The two cores of the normal liver tissue from 12 patients with metastatic colon carcinoma of the liver were included in each array block, as controls. Each tissue microarray block contained up to 60 tissue cores.
2. ImmunohistochemistryThe tissue microarray block was cut into 4-µm sections and immunohistochemical staining was performed as previously described [16]. The sections were subjected to antigen retrieval in 0.01 mol/L citrate buffer at pH 6.0 in a pressure cooker for 30 minutes. The sections were then incubated with a mouse monoclonal antibody to PEG10 (1:400, clone 4C10A7, MAB10294, Abnova Co., Taipei, Taiwan) for 30 minutes at room temperature. A negative control was obtained by replacing the primary antibody with isotype-matched irrelevant antibody. In order to validate the concordance between tissue microarrays and whole tumor sections, we further detected PEG10 expression for 40 corresponding whole tumor sections randomly chosen from the 218 cases.
PEG10 expression was assessed by two independent pathologists (C.-K.P. and H. B.), who were blinded to the clinicopathologic data, and any discrepancies were resolved by a consensus. A nearly homogeneous cytoplasmic immunostaining, with moderate staining intensity, was observed. For determining PEG10 expression, the proportion of stained tumor cells was semi-quantitatively assessed and each sample was scored on a scale of 0 to 4 (0, < 1%; 1, 1%-25%; 2, 26%-50%; 3, 51%-75%; 4, > 75%). Duplicate tissue cores for each tumor showed high levels of homogeneity for the proportion of stained cells. When there were differences between the duplicate tissue cores, the higher score was taken.
3. Statistical analysisAll statistical analyses were performed using SPSS ver. 18 statistical software (SPSS Inc., Chicago, IL). The association between PEG10 expression and clinicopathologic variables was analyzed using the chi-square test or Fisher exact test. The logistic regression analysis was used to predict tumor recurrence. Survival analysis was performed using the Kaplan-Meier method, and the difference in survival rates was assessed by the log-rank test. The Cox proportional hazards regression model was used to identify factors that were independently associated with survival. Significant prognostic factors identified by univariate analysis were entered into a multivariate analysis. A p-value of < 0.05 was considered to be statistically significant.
Results1. Clinicopathologic features of patientsThe mean patient age was 51.9 years (range, 17 to 76 years), 179 patients were men and 39 women. One hundred and seventy-one patients (78.4%) were infected with hepatitis B virus and 20 (9.2%) with hepatitis C virus. No viral marker was recognized in 27 patients (12.4%). One hundred and fifty-seven patients (72.0%) had suffered from tumor recurrence, 122 patients (56.0%) from early recurrence, and 35 patients (16.1%) from late recurrence. Eighty-nine patients (40.8%) died of HCC. Sixteen of the 105 deaths in this study were due to non-HCC causes. Ten of the 16 deaths were due to hepatic failure; five due to non-hepatic causes, and one due to unknown cause.
2. PEG10 protein expression in HCCImmunohistochemistry revealed that immunoreactivity for PEG10 was localized only in the cytoplasm of the tumor cells with moderate staining intensity. No immunoreactivity was found in normal hepatocytes (Fig. 1). We regarded PEG10 as positive when ≥ 1% of the tumor cells showed cytoplasmic immunoreactivity (Fig. 2). PEG10 protein expression was observed in 148 of the 218 HCCs (67.9%). PEG10 expression was significantly correlated with younger age (p=0.046), female (p=0.037), higher Edmondson grade (p=0.021), microvascular invasion (p=0.001), intrahepatic metastasis (p=0.008), higher AJCC T-stage (p=0.011), and higher α-fetoprotein level (p < 0.001). PEG10 expression was associated with early recurrence (p < 0.001), but not with late recurrence (p=0.863) (Table 1).
3. Prediction of early recurrence in HCCUnivariate analyses revealed that early recurrence was significantly correlated with larger tumor size (p=0.016), microvascular invasion (p < 0.001), intrahepatic metastasis (p < 0.001), higher AJCC T-stage (p < 0.001), higher BCLC stage (p < 0.001), lower albumin level (p=0.038), higher α-fetoprotein level (p=0.045), viral etiology (p < 0.001), and PEG10 expression (p < 0.001). Since AJCC T-stage and BCLC stage were associated with vascular invasion, we did not perform multiple analyses with these variables to avoid potential bias. An evaluation of the significant weight of the serum α-fetoprotein level was not performed due to missing data (n=178). On multivariate analyses, intrahepatic metastasis (p=0.005), viral etiology (p=0.004), and PEG10 expression (p=0.013) were independent predictors of early recurrence (Table 2).
4. Correlation between PEG10 expression and clinical outcomeThe 3-, 5-, 7-, and 9-year RFS and the overall survival (OS) rates for 218 HCC patients were 38.9%, 34.6%, 28.3%, 27.6% and 74.6%, 65.2%, 56.4%, 50.2%, respectively. Univariate analyses revealed that larger tumor size, Edmondson grade III, microvascular invasion, major portal vein invasion, intrahepatic metastasis, higher AJCC T-stage, higher BCLC stage, and lower albumin level showed unfavorable influences on both RFS and OS. Higher α-fetoprotein level and viral etiology showed unfavorable influences on RFS. PEG10 expression showed an unfavorable influence on RFS (p < 0.001) (Table 3). The 5-year RFS rate of the PEG10-positive group was significantly lower than that of the PEG10-negative group (25.4% vs. 53.7%) (Fig. 3A). The mean RFS of the PEG10-positive group and PEG10-negative group were 40.6 and 76.9 months, respectively. A subgroup analysis showed that among patients with α-fetoprotein ≤ 20 ng/mL (n=80), the PEG10-positive group (n=36) also showed an unfavorable influence on RFS (p=0.002) (Fig. 3B). Further analysis revealed that among patients with tumor size ≤ 5.0 cm (n=134) and patients at AJCC T-stage 1 (n=91), the PEG10-positive groups (n=90 and n=51, respectively) tended to show unfavorable influences on RFS (p=0.072 and p=0.079, respectively) (Fig. 3C and D).
PEG10 expression showed an unfavorable influence on OS (p=0.007) (Table 3). The 5-year OS rate of the PEG10-positive group was significantly lower than that of the PEG10-negative group (59.7% vs. 76.8%) (Fig. 4). Mean OS of the PEG10-positive group and PEG10-negative group were 88.1 and 112.7 months, respectively.
On multivariate analyses, intrahepatic metastasis and lower albumin level were found to be independent predictors of both shorter RFS and shorter OS. Microvascular invasion and viral etiology were found to be independent predictors of shorter RFS. PEG10 expression (p=0.005) was found to be an independent predictor of shorter RFS, but not of OS (p=0.128). PEG10-positive patients were more likely to suffer from recurrence than PEG10-negative patients (hazard ratio, 1.724) (Table 4).
DiscussionPEG10 might be involved in oncogenesis by increasing the apoptotic resistance of the tumor cells. In HCC, PEG10 decreased cell death through interaction with seven in absentia homolog 1, which is a mediator of apoptosis [17]. Moreover, PEG10 knockdown inhibited the proliferation of the HepG2 HCC cells [5], while targeted disruption of the mouse PEG10 gene caused early embryonic lethality due to defects in the placenta [18]. Recent reports demonstrated that PEG10 expression can be regulated by proto-oncogene c-MYC [5], by E2F transcription factors that modulate the cell cycle [19], and by the sex hormone androgen [20]. The exact mechanism by which PEG10 signals is unclear, but it is known to interact with tumor growth factor β type I receptor ALK1 [21].
Although PEG10 overexpression in HCC was previously reported [7-10], the prognostic significance of PEG10 protein expression in HCC has not yet been reported. In this study, we found that PEG10 expression was significantly associated with higher Edmondson grade, microvascular invasion, intrahepatic metastasis, higher AJCC T-stage, and higher αfetoprotein level. Moreover, PEG10 expression was an independent predictor of early recurrence and shorter RFS. PEG10 expression could be used as a tool for early detection of HCC recurrence.
Measurement of the serum α-fetoprotein level is widely used for routine surveillance and noninvasive diagnosis of HCC, as well as to evaluate the prognosis and monitor recurrence following treatment [22]. As a limitation, elevated serum α-fetoprotein is observed in only 60% to 70% of HCC patients and, to a lesser extent (33%-65%), in patients with smaller HCCs [23]. Cucchetti et al. [24] asserted that preoperative high serum α-fetoprotein level (> 60 ng/mL) could be a risk factor for recurrence after resection in patients with liver cirrhosis. However, there is no effective marker to monitor recurrence and guide treatment for α-fetoprotein– negative (≤ 20 ng/mL) patients after hepatectomy. In this study, tumor recurrence was detected in 51 of 80 α-fetoprotein–negative patients (63.8%). We found that within α-fetoprotein–negative patients, the PEG10-positive group still showed an unfavorable influence on RFS (p=0.002). PEG10 might be a useful biomarker to monitor recurrence for α-fetoprotein–negative HCC patients.
HCC patients with tumor size ≤ 5.0 cm and patients at AJCC T-stage 1 are generally considered to have relatively low risk for recurrence [25]; however, in this study, 91 of 134 (67.9%) with ≤ 5.0 cm and 53 of 91 (58.2%) AJCC T-stage 1 showed recurrence. Within these populations, we found that the PEG10-positive groups tended to show unfavorable influences on RFS (p=0.072 and p=0.079, respectively).
ConclusionOur findings indicate, for the first time, that PEG10 protein could be a potential biomarker predicting early recurrence and RFS in HCC patients after curative resection, even in those with normal serum α-fetoprotein levels. These findings also indicate possible new lines of research for the development of therapeutic approaches targeting PEG10. Further work is needed to explore functional roles of the PEG10 gene, which might play in the development and progression of HCC.
Fig. 1.Immunostaining of paternally expressed gene 10 in normal liver shows no immunoreactivity in normal hepatocytes (horseradish peroxidase staining, ×200). 
Fig. 2.Immunostaining of paternally expressed gene 10 in hepatocellular carcinomas showing positive cytoplasmic expression (A) or negative expression (B) (horseradish peroxidase staining, ×200). 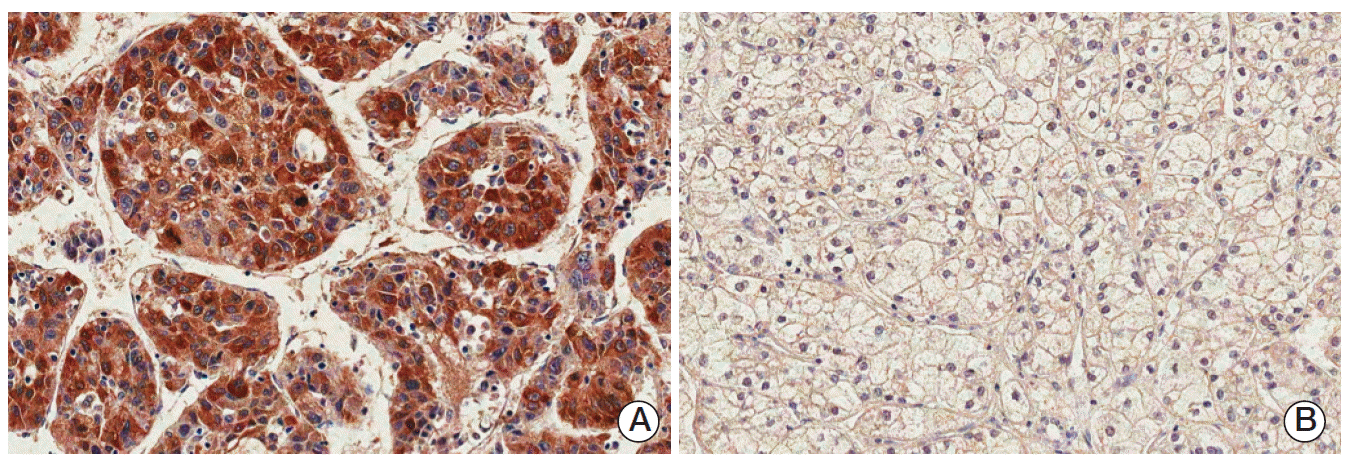
Fig. 3.Kaplan-Meier survival curves showing recurrence-free survival among all patients (A), patients with α-fetoprotein (AFP) ≤ 20 ng/mL (B), patients with tumor size ≤ 5.0 cm (C), and patients at American Joint Committee on Cancer (AJCC) T-stage 1 (D) according to the paternally expressed gene 10 (PEG10) expression. 
Fig. 4.Kaplan-Meier survival curves showing overall survival for paternally expressed gene 10 (PEG10) expression in 218 hepatocellular carcinomas. 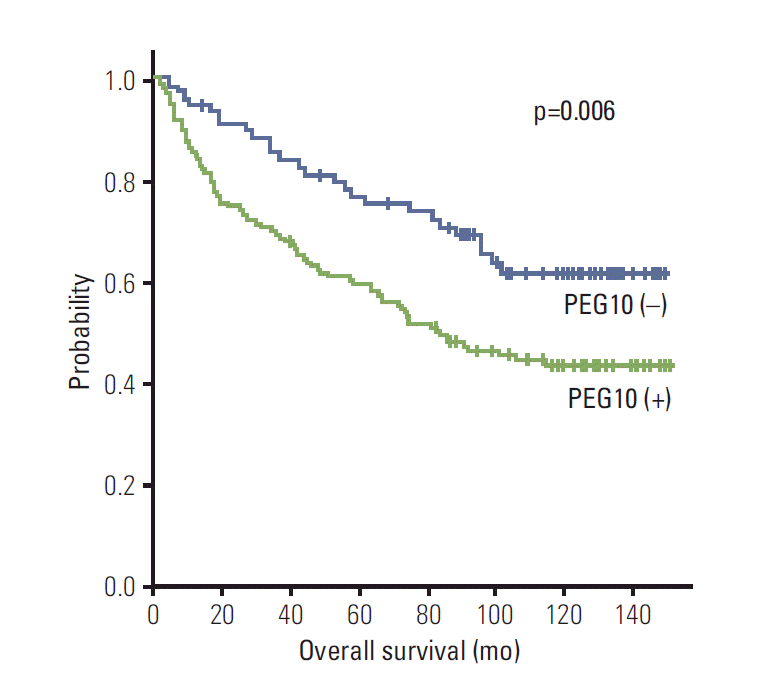
Table 1.Correlation between PEG10 expression and the clinicopathologic features in 218 hepatocellular carcinomas
Table 2.Univariate and multivariate logistic regression models for predicting early tumor recurrence in 183 hepatocellular carcinomas
Table 3.Univariate analyses of recurrence-free survival and overall survival in 218 hepatocellular carcinomas
Table 4.Multivariate analyses of recurrence-free survival and overall survival in 218 hepatocellular carcinomas References1. Regimbeau JM, Abdalla EK, Vauthey JN, Lauwers GY, Durand F, Nagorney DM, et al. Risk factors for early death due to recurrence after liver resection for hepatocellular carcinoma: results of a multicenter study. J Surg Oncol. 2004;85:36–41.
2. Simonetti RG, Liberati A, Angiolini C, Pagliaro L. Treatment of hepatocellular carcinoma: a systematic review of randomized controlled trials. Ann Oncol. 1997;8:117–36.
3. Dobashi Y, Goto A, Endo T, Ooi A. Genetic aberrations as the targets of oncology research: involvement of paraffin-embedded tissues. Histol Histopathol. 2014;29:191–205.
4. Ono R, Kobayashi S, Wagatsuma H, Aisaka K, Kohda T, Kaneko-Ishino T, et al. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics. 2001;73:232–7.
5. Li CM, Margolin AA, Salas M, Memeo L, Mansukhani M, Hibshoosh H, et al. PEG10 is a c-MYC target gene in cancer cells. Cancer Res. 2006;66:665–72.
6. Liu DC, Yang ZL, Jiang S. Identification of PEG10 and TSG101 as carcinogenesis, progression, and poor-prognosis related biomarkers for gallbladder adenocarcinoma. Pathol Oncol Res. 2011;17:859–66.
7. Tsou AP, Chuang YC, Su JY, Yang CW, Liao YL, Liu WK, et al. Overexpression of a novel imprinted gene, PEG10, in human hepatocellular carcinoma and in regenerating mouse livers. J Biomed Sci. 2003;10:625–35.
8. Ip WK, Lai PB, Wong NL, Sy SM, Beheshti B, Squire JA, et al. Identification of PEG10 as a progression related biomarker for hepatocellular carcinoma. Cancer Lett. 2007;250:284–91.
9. Tsuji K, Yasui K, Gen Y, Endo M, Dohi O, Zen K, et al. PEG10 is a probable target for the amplification at 7q21 detected in hepatocellular carcinoma. Cancer Genet Cytogenet. 2010;198:118–25.
10. Dong H, Zhang H, Liang J, Yan H, Chen Y, Shen Y, et al. Digital karyotyping reveals probable target genes at 7q21.3 locus in hepatocellular carcinoma. BMC Med Genomics. 2011;4:60.
11. Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:468–503.
12. Kumada T, Nakano S, Takeda I, Sugiyama K, Osada T, Kiriyama S, et al. Patterns of recurrence after initial treatment in patients with small hepatocellular carcinoma. Hepatology. 1997;25:87–92.
13. Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC cancer staging manual. 7th edNew York: Springer; 2010. p. 191–5.
14. Llovet JM, Bru C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329–38.
15. Imamura H, Matsuyama Y, Tanaka E, Ohkubo T, Hasegawa K, Miyagawa S, et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol. 2003;38:200–7.
16. Ahn S, Hyeon J, Park CK. Metadherin is a prognostic predictor of hepatocellular carcinoma after curative hepatectomy. Gut Liver. 2013;7:206–12.
17. Okabe H, Satoh S, Furukawa Y, Kato T, Hasegawa S, Nakajima Y, et al. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 2003;63:3043–8.
18. Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. 2006;38:101–6.
19. Wang C, Xiao Y, Hu Z, Chen Y, Liu N, Hu G. PEG10 directly regulated by E2Fs might have a role in the development of hepatocellular carcinoma. FEBS Lett. 2008;582:2793–8.
20. Jie X, Lang C, Jian Q, Chaoqun L, Dehua Y, Yi S, et al. Androgen activates PEG10 to promote carcinogenesis in hepatic cancer cells. Oncogene. 2007;26:5741–51.
21. Lux A, Beil C, Majety M, Barron S, Gallione CJ, Kuhn HM, et al. Human retroviral gag- and gag-pol-like proteins interact with the transforming growth factor-beta receptor activin receptor-like kinase 1. J Biol Chem. 2005;280:8482–93.
22. Johnson PJ. The role of serum alpha-fetoprotein estimation in the diagnosis and management of hepatocellular carcinoma. Clin Liver Dis. 2001;5:145–59.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||